Short-Term Local Adaptation of Historical Common Bean (Phaseolus vulgaris L.) Varieties and Implications for In Situ Management of Bean Diversity
Abstract
:1. Introduction
2. Results
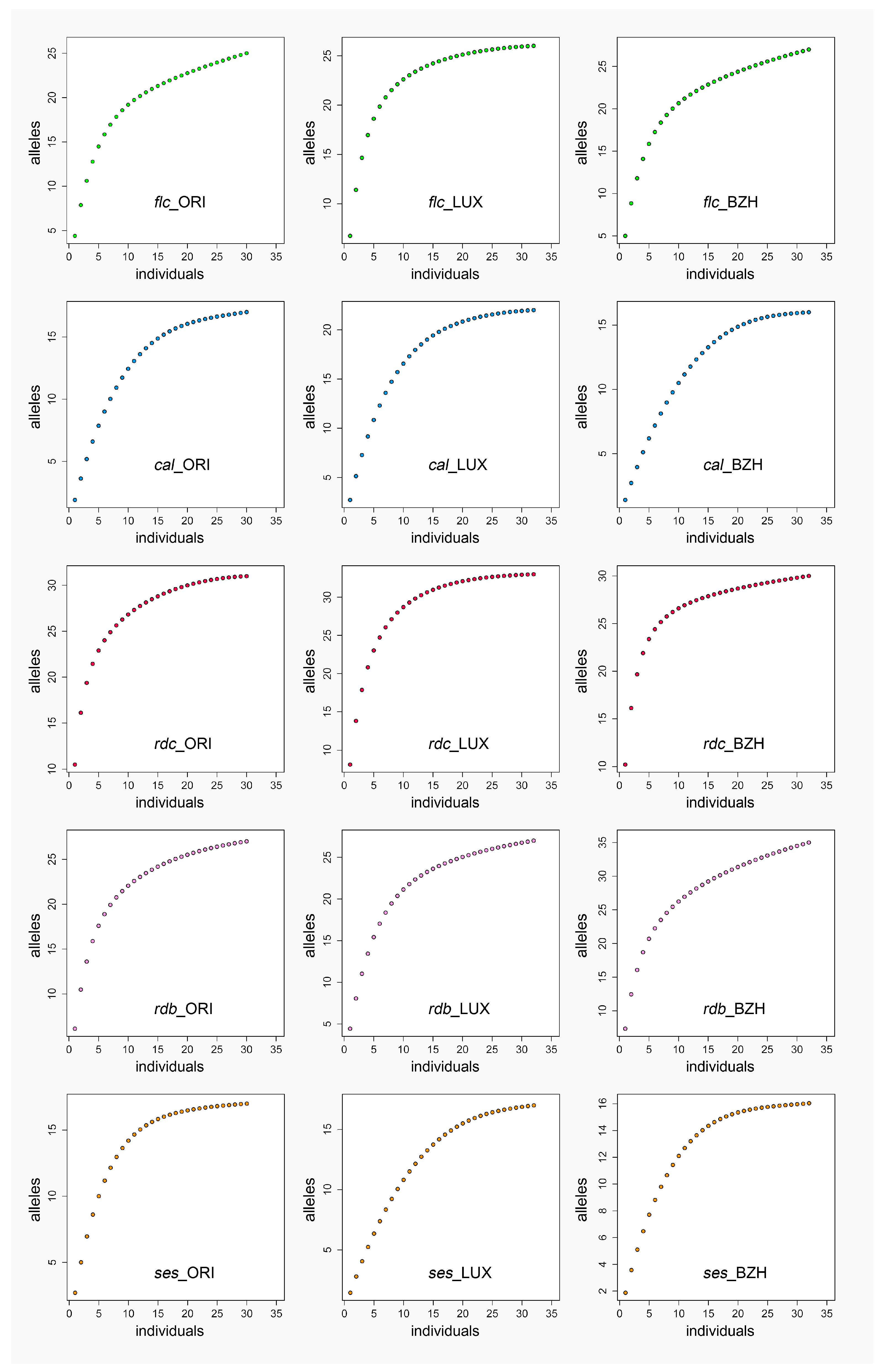
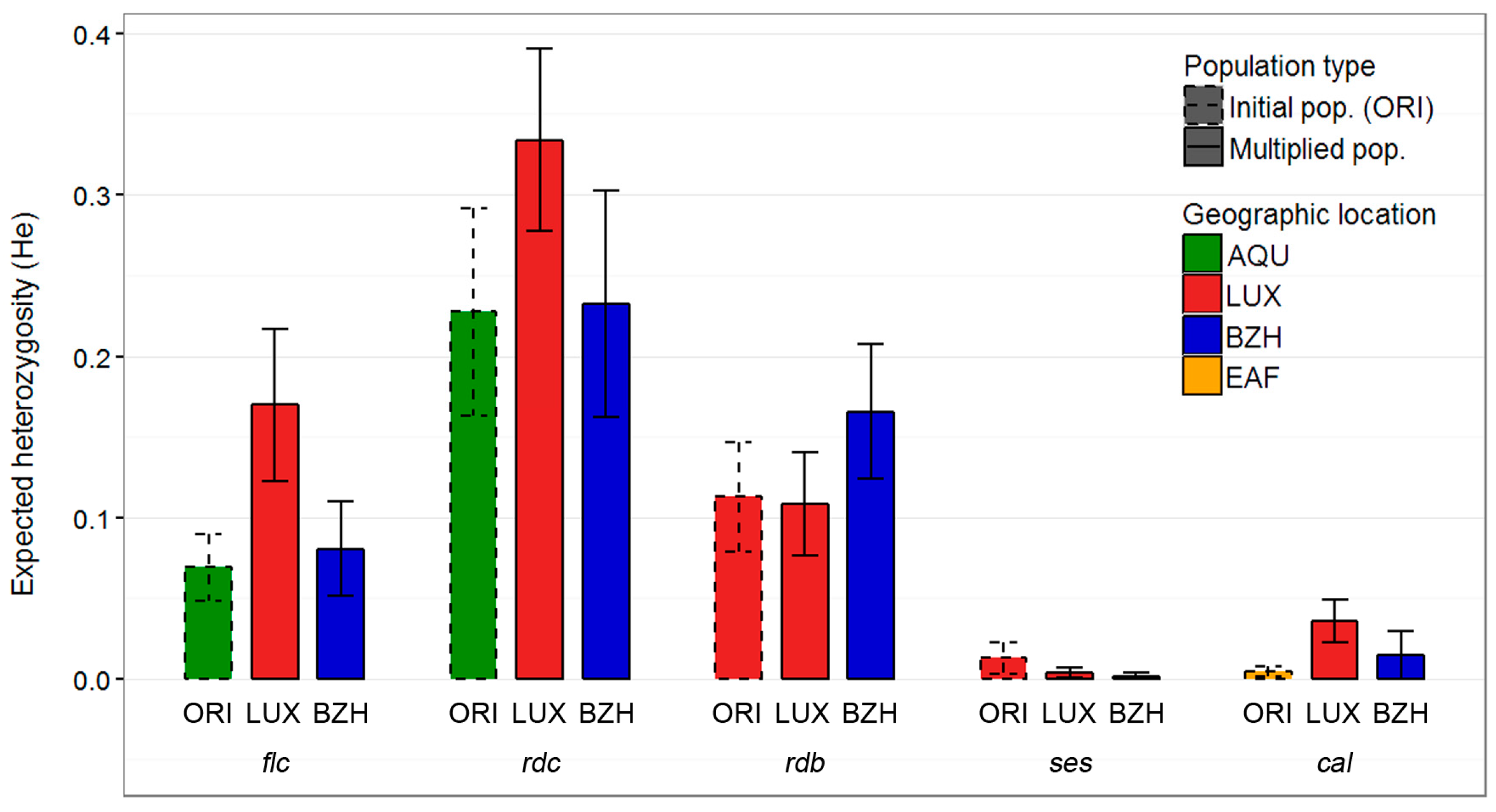
2.1. Genetic Diversity
2.2. Genetic Differentiation
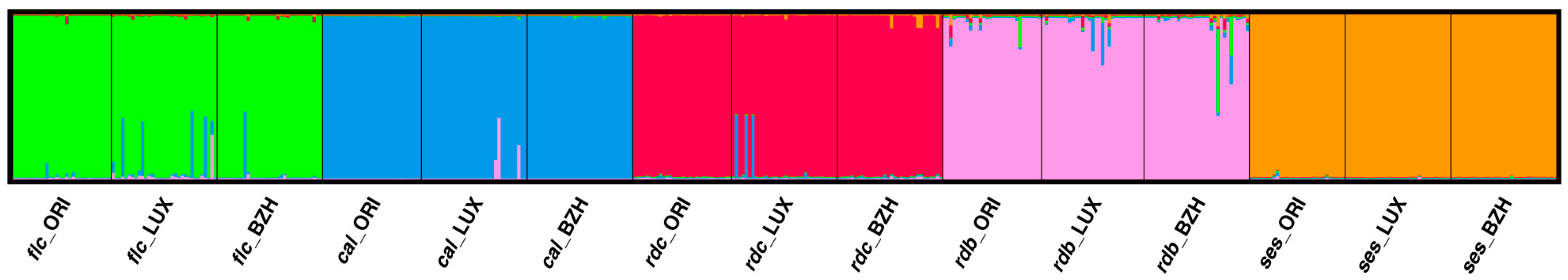
2.3. Population Genetic Structure Analysis
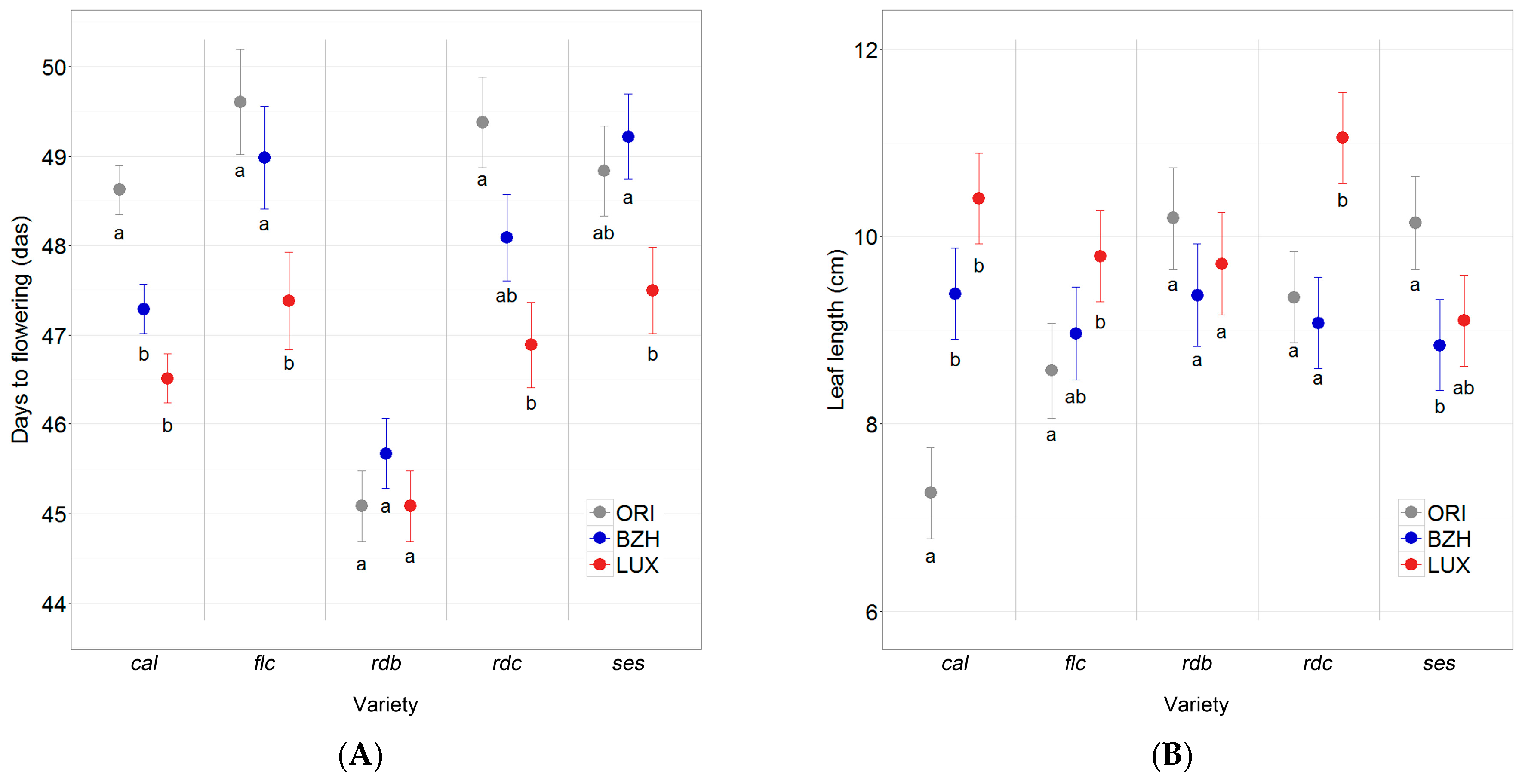
2.4. Phenotypic Differentiation
3. Discussion
3.1. Genetic Diversity
3.2. Genetic Differentiation
3.3. Genetic Relationships
3.4. Phenotypic Differentiation Indicates Local Adaptation
3.5. Implications for the In Situ Management of Bean Diversity in Fields and Gardens
4. Materials and Methods
4.1. Experimental Setup
4.1.1. Varieties
4.1.2. Experimental Design
4.1.3. Sampling for DNA Extraction and Phenotypic Observations
4.2. Plant DNA Extraction and SSR Genotyping
4.3. SSR Data Analysis
4.4. Observation of Phenotypic Traits
4.5. Analysis of Phenotypic Data
4.6. Semi-Directive Interviews
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| SSR | Simple sequence repeat/microsatellite |
| TSW | 1000-seed weight |
| cal | ‘Calima’ |
| flc | ‘Flageolet Chevrier’ |
| rdb | ‘Roi des Belges’ |
| rdc | ‘Rognon de Coq’ |
| ses | ‘Saint Esprit à œil rouge’ |
| AQU | Aquitaine |
| BZH | Brittany |
| LUX | Luxembourg |
| ORI | Original seed lot employed to establish the field trial |
| AMOVA | Analysis of Molecular Variance |
| He | Expected heterozygosity |
| PCoA | Principal coordinate analysis |
| FST | Fixation index |
| das | Days after sowing |
| HBB | Halo bacterial blight |
| Na | Number of alleles |
| MCMC | Markov Chain Monte Carlo |
| HSD | Tukey’s Honestly Significant Difference |
| pd | Relative effect |
References
- Singh, S.P.; Gutiérrez, J.A.; Molina, A.; Urrea, C.; Gepts, P. Genetic Diversity in Cultivated Common Bean: II. Marker-Based Analysis of Morphological and Agronomic Traits. Crop Sci. 1991, 31, 23. [Google Scholar] [CrossRef]
- Kwak, M.; Gepts, P. Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae). Theor. Appl. Genet. 2009, 118, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Gepts, P.; Kmiecik, K.; Pereira, P.; Bliss, F.A. Dissemination pathways of common bean (Phaseolus vulgaris, Fabaceae) deduced from phaseolin electrophoretic variability. I. The Americas. Econ. Bot. 1988, 42, 73–85. [Google Scholar] [CrossRef]
- Zewen, A.C. The introduction of the common bean (Phaseolus vulgaris L.) into Western Europe and the phenotypic variation of dry beans collected in The Netherlands in 1946. Euphytica 1997, 94, 319–328. [Google Scholar]
- Maras, M.; Šuštar-Vozlič, J.; Kainz, W.; Meglič, V. Genetic Diversity and Dissemination Pathways of Common Bean in Central Europe. J. Am. Soc. Hortic. Sci. 2013, 138, 297–305. [Google Scholar]
- Gioia, T.; Logozzo, G.; Attene, G.; Bellucci, E.; Benedettelli, S.; Negri, V.; Papa, R.; Spagnoletti Zeuli, P. Evidence for Introduction Bottleneck and Extensive Inter-Gene Pool (Mesoamerica × Andes) Hybridization in the European Common Bean (Phaseolus vulgaris L.) Germplasm. PLoS ONE 2013, 8, e75974. [Google Scholar] [CrossRef] [PubMed]
- Angioi, S.A.; Rau, D.; Attene, G.; Nanni, L.; Bellucci, E.; Logozzo, G.; Negri, V.; Spagnoletti Zeuli, P.L.; Papa, R. Beans in Europe: Origin and structure of the European landraces of Phaseolus vulgaris L. Theor. Appl. Genet. 2010, 121, 829–843. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations Production and Trade Data. Available online: http://faostat3.fao.org/home/E (accessed on 6 May 2016).
- European Commission International Trade Statistics. Available online: http://ec.europa.eu/eurostat/web/international-trade/data/database (accessed on 5 June 2016).
- Unilet. Union Nationale Interprofessionnelle des Légumes Transformés; Unilet Infos: Paris, France, 2015; pp. 28–31. [Google Scholar]
- Kastler, G. Les Semences Paysannes: Situation Actuelle, Difficultés Techniques, Besoin d’un Cadre Juridique. Les Dossiers de l'environnement de l'INRA 2006, 30, 53–56. (In Franch) [Google Scholar]
- Bocci, R.; Chable, V. Peasant seeds in Europe: Stakes and prospects. J. Agric. Environ. Int. Dev. 2009. [Google Scholar] [CrossRef]
- Brouwer, B.O.; Murphy, K.M.; Jones, S.S. Plant breeding for local food systems: A contextual review of end-use selection for small grains and dry beans in Western Washington. Renew. Agric. Food Syst. 2015, 31, 1–13. [Google Scholar]
- Sánchez, E.; Sifres, A.; Casañas, F.; Nuez, F. Common bean (Phaseolus vulgaris L.) landraces in Catalonia, a Mesoamerican germplasm hotspot to be preserved. J. Hortic. Sci. Biotechnol. 2007, 82, 529–534. [Google Scholar] [CrossRef]
- Thomas, M.; Demeulenaere, E.; Dawson, J.C.; Khan, A.R.; Galic, N.; Jouanne-Pin, S.; Remoue, C.; Bonneuil, C.; Goldringer, I. On-farm dynamic management of genetic diversity: The impact of seed diffusions and seed saving practices on a population-variety of bread wheat: On-farm crop metapopulation of bread wheat. Evol. Appl. 2012, 5, 779–795. [Google Scholar] [CrossRef] [PubMed]
- Morgante, M.; Oliveiri, M. PCR-amplified microsatellites as markers in plant genetics. Plant J. 1993, 3, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Park, S.J.; Poysa, V. Abundance and variation of microsatellite DNA sequences in beans (Phaseolus and Vigna). Genome 1999, 42, 27–34. [Google Scholar] [CrossRef]
- Gaitán-Solís, E.; Duque, M.C.; Edwards, K.J.; Tohme, J. Microsatellite Repeats in Common Bean (Phaseolus vulgaris). Crop Sci. 2002, 42, 2128–2136. [Google Scholar] [CrossRef]
- Blair, M.W.; Torres, M.M.; Pedraza, F.; Giraldo, M.C.; Buendía, H.F.; Hurtado, N. Development of microsatellite markers for common bean (Phaseolus vulgaris L.) based on screening of non-enriched, small-insert genomic libraries. Genome 2009, 52, 772–782. [Google Scholar] [PubMed]
- Córdoba, J.M.; Chavarro, C.; Rojas, F.; Muñoz, C.; Blair, M.W. Identification and Mapping of Simple Sequence Repeat Markers from Common Bean (Phaseolus vulgaris L.) Bacterial Artificial Chromosome End Sequences for Genome Characterization and Genetic–Physical Map Integration. Plant Genome J. 2010, 3, 154. [Google Scholar] [CrossRef]
- Blair, M.W.; Pedraza, F.; Buendia, H.F.; Gaitán-Solís, E.; Beebe, S.E.; Gepts, P.; Tohme, J. Development of a genome-wide anchored microsatellite map for common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2003, 107, 1362–1374. [Google Scholar] [CrossRef] [PubMed]
- Blair, M.W.; Díaz, L.M.; Buendía, H.F.; Duque, M.C. Genetic diversity, seed size associations and population structure of a core collection of common beans (Phaseolus vulgaris L.). Theor. Appl. Genet. 2009, 119, 955–972. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Thépot, S.; Galic, N.; Jouanne-Pin, S.; Remoué, C.; Goldringer, I. Diversifying mechanisms in the on-farm evolution of crop mixtures. Mol. Ecol. 2015, 24, 2937–2954. [Google Scholar] [CrossRef] [PubMed]
- Raggi, L.; Ceccarelli, S.; Negri, V. Evolution of a barley composite cross-derived population: An insight gained by molecular markers. J. Agric. Sci. 2016, 154, 23–39. [Google Scholar] [CrossRef]
- Tiranti, B.; Negri, V. Selective microenvironmental effects play a role in shaping genetic diversity and structure in a Phaseolus vulgaris L. landrace: Implications for on-farm conservation. Mol. Ecol. 2007, 16, 4942–4955. [Google Scholar] [CrossRef] [PubMed]
- Serpolay, E.; Goldringer, I.; Negri, V.; Raggi, L.; Tissi, C.; Chable, V. On-farm breeding of bean populations, an approach of genetic diversity evolution. In Shaping the Future of Agriculture: The Role of Diversity in Lowinput and Organic Cropping Systems, Proceedings of the SOLIBAM 1st Stakeholder Congress, Rome, Italy, 19–20 April 2012; Grottaferrata: Rome, Italy, 2012. [Google Scholar]
- Horneburg, B.; Becker, H.C. Crop Adaptation in On-Farm Management by Natural and Conscious Selection: A Case Study with Lentil. Crop Sci. 2008, 48, 203. [Google Scholar] [CrossRef]
- Dawson, J.C.; Serpolay, E.; Giuliano, S.; Schermann, N.; Galic, N.; Berthellot, J.-F.; Chesneau, V.; Ferté, H.; Mercier, F.; Osman, A.; et al. Phenotypic diversity and evolution of farmer varieties of bread wheat on organic farms in Europe. Genet. Resour. Crop Evol. 2013, 60, 145–163. [Google Scholar] [CrossRef]
- Rhoné, B.; Vitalis, R.; Goldringer, I.; Bonnin, I. Evolution of flowering time in experimental wheat populations: A comprehensive approach to detect genetic signatures of natural selection. Evolution 2010, 64, 2110–2115. [Google Scholar] [CrossRef] [PubMed]
- Bitocchi, E.; Bellucci, E.; Giardini, A.; Rau, D.; Rodriguez, M.; Biagetti, E.; Santilocchi, R.; Spagnoletti Zeuli, P.; Gioia, T.; Logozzo, G.; et al. Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol. 2013, 197, 300–313. [Google Scholar] [CrossRef] [PubMed]
- Allard, R.W. The mating system and microevolution. Genetics 1975, 79, 115–126. [Google Scholar] [PubMed]
- Ibarra-Perez, F.J.; Ehdaie, B.; Waines, J.G. Estimation of Outcrossing Rate in Common Bean. Crop Sci. 1997, 37, 60–65. [Google Scholar] [CrossRef]
- Martins, G.B.; Adams, M.W. Landraces of Phaseolus vulgaris (Fabaceae) in Northern Malawi. II. Generation and maintenance of variability. Econ. Bot. 1987, 204–215. [Google Scholar] [CrossRef]
- Serpolay, E.; Schermann, N.; Dawson, J.; van Bueren, E.T.L.; Goldringer, I.; Chable, V. Phenotypic Changes in Different Spinach Varieties Grown and Selected under Organic Conditions. Sustainability 2011, 3, 1616–1636. [Google Scholar] [CrossRef]
- Serpolay-Besson, E.; Giuliano, S.; Schermann, N.; Chable, V. Evaluation of Evolution and Diversity of Maize Open-Pollinated Varieties Cultivated under Contrasted Environmental and Farmers’ Selection Pressures: A Phenotypical Approach. Open J. Genet. 2014, 4, 125–145. [Google Scholar] [CrossRef]
- Joshi, J.; Schmid, B.; Caldeira, M.C.; Dimitrakopoulos, P.G.; Good, J.; Harris, R.; Hector, A.; Huss-Danell, K.; Jumpponen, A.; Minns, A.; et al. Local adaptation enhances performance of common plant species. Ecol. Lett. 2001, 4, 536–544. [Google Scholar] [CrossRef]
- Leimu, R.; Fischer, M. A Meta-Analysis of Local Adaptation in Plants. PLoS ONE 2008, 3, e4010. [Google Scholar] [CrossRef] [PubMed]
- Dudley, S.A. The Response to Differing Selection on Plant Physiological Traits: Evidence for Local Adaptation. Evolution 1996, 50, 103. [Google Scholar] [CrossRef]
- Roach, D.A.; Wulff, R.D. Maternal effects in plants. Annu. Rev. Ecol. Syst. 1987, 18, 209–235. [Google Scholar] [CrossRef]
- Lacey, E.R. What is an adaptive environmentally induced parental effect? In Maternal Effects as Adaptations; Mousseau, T.A., Fox, C.W., Eds.; Oxford University Press: New York, NY, USA, 1998. [Google Scholar]
- Galloway, L.F. Maternal effects provide phenotypic adaptation to local environmental conditions: Research review. New Phytol. 2005, 166, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Sadras, V.O. Evolutionary aspects of the trade-off between seed size and number in crops. Field Crops Res. 2007, 100, 125–138. [Google Scholar] [CrossRef]
- Klaedtke, S.; Jacques, M.-A.; Raggi, L.; Préveaux, A.; Bonneau, S.; Negri, V.; Chable, V.; Barret, M. Terroir is a key driver of seed-associated microbial assemblages. Environ. Microbiol. 2016, 18, 1792–1804. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.M.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.H.M.; et al. Deciphering the Rhizosphere Microbiome for Disease-Suppressive Bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, A.; Bakker, M.G.; Badri, D.V.; Manter, D.K.; Vivanco, J.M. Relationships between Arabidopsis genotype-specific biomass accumulation and associated soil microbial communities. Botany 2013, 91, 123–126. [Google Scholar] [CrossRef]
- Panke-Buisse, K.; Poole, A.C.; Goodrich, J.K.; Ley, R.E.; Kao-Kniffin, J. Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J. 2014, 9, 980–989. [Google Scholar] [CrossRef] [PubMed]
- Barret, M.; Guimbaud, J.-F.; Darrasse, A.; Jacques, M.-A. Plant microbiota affects seed transmission of phytopathogenic microorganisms. Mol. Plant Pathol. 2016, 17, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M. Gestion Dynamique à la Ferme de L’agrobiodiversité: Relation Entre la Structure des Populations de blé Tendre et les Pratiques Humaines; Université Paris Diderot, Paris 7: Paris, France, 2011. [Google Scholar]
- Negri, V.; Tiranti, B. Effectiveness of in situ and ex situ conservation of crop diversity. What a Phaseolus vulgaris L. landrace case study can tell us. Genetica 2010, 138, 985–998. [Google Scholar] [CrossRef] [PubMed]
- Nelson, G.C.; Rosegrant, M.W.; Koo, J.; Robertson, R.; Sulser, T.; Zhu, T.; Ringler, C.; Msangi, S.; Palazzo, A.; Batka, M.; et al. Climate Change: Impact on Agriculture and Costs of Adaptation; International Food Policy Research Institute, Ed.; Food Policy Report; International Food Policy Research Institute: Washington, DC, USA, 2009. [Google Scholar]
- Enjalbert, J.; Dawson, J.C.; Paillard, S.; Rhoné, B.; Rousselle, Y.; Thomas, M.; Goldringer, I. Dynamic management of crop diversity: From an experimental approach to on-farm conservation. C. R. Biol. 2011, 334, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Newton, A.C.; Akar, T.; Baresel, J.P.; Bebeli, P.J.; Bettencourt, E.; Bladenopoulos, K.V.; Czembor, J.H.; Fasoula, D.A.; Katsiotis, A.; Koutis, K.; et al. Cereal landraces for sustainable agriculture: A review. Agron. Sustain. Dev. 2010, 30, 237–269. [Google Scholar]
- Soliman, K.M.; Allard, R.W. Grain Yield of Composite Cross Populations of Barley: Effects of Natural Selection. Crop Sci. 1991, 31, 705–708. [Google Scholar] [CrossRef]
- Soleri, D.; Smith, S.E. Morphological and Phenological Comparisons of Two Hopi Maize Varieties Conserved in situ and ex situ. Econ. Bot. 1995, 49, 56–77. [Google Scholar] [CrossRef]
- Rice, E.; Smale, M.; Blanco, J.-L. Farmers’ Use of Improved Seed Selection Practices in Mexican Maize: Evidence and Issues from the Sierra de Santa Marta. World Dev. 1998, 26, 165–1640. [Google Scholar] [CrossRef]
- Tin, H.; Berg, T.; Bjørnstad, Å. Diversity and adaptation in rice varieties under static (ex situ) and dynamic (in situ) management. Euphytica 2001, 122, 491–502. [Google Scholar] [CrossRef]
- Gómez, O.J.; Blair, M.W.; Frankow-Lindberg, B.E.; Gullberg, U. Comparative Study of Common Bean (Phaseolus vulgaris L.) Landraces Conserved ex situ in Genebanks and in situ by Farmers. Genet. Resour. Crop Evol. 2005, 52, 371–380. [Google Scholar] [CrossRef]
- Bradshaw, A.D. Ecological significance of genetic variation between populations. In Perspectives on Plant Population Ecology; Dirzo, R., Sarukhan, J., Eds.; Sinauer Associates, Inc.: Sunderland, MA, USA, 1984; pp. 213–228. [Google Scholar]
- Gallagher, J.L.; Somers, G.F.; Grant, D.M.; Seliskar, D.M. Persistent Differences in Two Forms of Spartina Alterniflora: A Common Garden Experiment. Ecology 1988, 69, 1005–1008. [Google Scholar] [CrossRef]
- Vilmorin-Andrieux et Cie. Description des Plantes Potagères; Vilmorin: Paris, France, 1855. [Google Scholar]
- Vilmorin-Andrieux, S.A. Dictionnaire Vilmorin des Plantes Potagères; Vilmorin: Paris, France, 1947. [Google Scholar]
- Raggi, L.; Tissi, C.; Mazzucato, A.; Negri, V. Molecular polymorphism related to flowering trait variation in a Phaseolus vulgaris L. collection. Plant Sci. 2014, 215–216, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, E.; Davis, S. The ARES Package. R Package Version 1.2-2. Available online: http://ftp.uni-bayreuth.de/math/statlib/R/CRAN/doc/packages/ARES.pdf (accessed on 1 February 2017).
- Peakall, R.; Smouse, P.E. genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- VIB/UGent Calculate and Draw Custom Venn Diagrams. Available online: http://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed on 12 December 2016).
- Conner, J.K.; Hartl, D.L. A Primer of Ecological Genetics; Sinauer Associates: Sunderland, MA, USA, 2004. [Google Scholar]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Gao, H.; Williamson, S.; Bustamante, C.D. An MCMC Approach for Joint Inference of Population Structure and Inbreeding Rates from Multi-Locus Genotype Data. Genetics 2007, 176, 1635–1651. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- International Board for Plant Genetic Resources (IPGRI). Phaseolus Vulgaris Descriptors; International Board for Plant Genetic Resources: Rome, Italy, 1982. [Google Scholar]
- Van Schoonhoven, A.; Pastor-Corrales, M.A.; Centro Internacional de Agricultura Tropical. Standard System for the Evaluation of Bean Germplasm; Centro Internacional de Agricultura Tropical: Cali, Colombia, 1987. [Google Scholar]
- Collmer, C.W.; Marston, M.F.; Taylor, J.C.; Jahn, M. The I Gene of Bean: A Dosage-Dependent Allele Conferring Extreme Resistance, Hypersensitive Resistance, or Spreading Vascular Necrosis in Response to the Potyvirus Bean common mosaic virus. Mol. Plant. Microbe Interact. 2000, 13, 1266–1270. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Development Core Team: Vienna, Austria, 2016. [Google Scholar]
- Pinheiro, J.C.; Bates, D.M.; DebRoy, S. R-Core Nlme: Linear and Nonlinear Mixed Effects Models. R Package Version 3.1-131. Available online: https://CRAN.R-project.org/package=nlme (accessed on 6 February 2017).
- Fang, L.; Loughin, T.M. Analyzing binomial data in a split-plot design: Classical approaches or modern techniques? In 16th Annual Conference Proceedings, Paper 16; Kansas State University Libraries, New Prairie Press: Hale, TX, USA, 2004. [Google Scholar]
- Lenth, R.V. Lsmeans: Least-squares means. J. Stat. Softw. 2016, 69. [Google Scholar] [CrossRef]
- Brunner, E.; Puri, M.L. Nonparametric methods in factorial designs. Stat. Pap. 2001, 42, 1–52. [Google Scholar] [CrossRef]
- Shah, D.A.; Madden, L.V. Nonparametric Analysis of Ordinal Data in Designed Factorial Experiments. Phytopathology 2004, 94, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Konietschke, F.; Friedrich, S.; Brunner, E.; Pauly, M. RankFD: Rank-Based Tests for General Factorial Designs. R package version 0.0.0.2. Available online: https://CRAN.R-project.org/package=rankFD (accessed on 28 June 2016).


| Variety | Site/Origin | Population (Code) | TSW (g) | Information on Original Seed Lot |
|---|---|---|---|---|
| Calima (cal) modern variety (control) | Original seed lot | cal_ORI | 250 | Harvested in East Africa in 2010 |
| Brittany | cal_BZH | 278 | ||
| Luxembourg | cal_LUX | 301 | ||
| Flageolet Chevrier (flc) historical variety | Original seed lot | flc_ORI | 235 | Harvested in AQU in 2011 (organic) |
| Brittany | flc_BZH | 244 | ||
| Luxembourg | flc_LUX | 256 | ||
| Rognon de Coq (rdc) historical variety | Original seed lot | rdc_ORI | 340 | Harvested in AQU in 2011 (organic) |
| Brittany | rdc_BZH | 415 | ||
| Luxembourg | rdc_LUX | 452 | ||
| Roi des Belges (rdb) historical variety | Original seed lot | rdb_ORI | 442 | Harvested in LUX in 2011 (organic) |
| Brittany | rdb_BZH | 320 | ||
| Luxembourg | rdb_LUX | 376 | ||
| St. Esprit à œil rouge (ses) historical variety | Original seed lot | ses_ORI | 884 | Harvested in LUX in 2011 (organic) |
| Brittany | ses_BZH | 682 | ||
| Luxembourg | ses_LUX | n.a. |
| Variety | ORI vs. BZH | ORI vs. LUX | BZH vs. LUX |
|---|---|---|---|
| “Flageolet Chevrier” ( flc) | 0.01 | 0.12 *** | 0.09 *** |
| “Rognon de Coq” ( rdc) | 0.03 | 0.19 *** | 0.20 *** |
| “Roi des Belges” ( rdb) | 0.01 | 0.00 | 0.00 |
| “St. Esprit” ( ses) | 0.07 | 0.06 | −0.02 |
| “Calima” ( cal) | 0.02 | 0.01 | 0.03 |
| Marker | Linkage Group | SSR Motif | Primer FOR | Primer REV | Predicted Size | References |
|---|---|---|---|---|---|---|
| BM200 | b01 | (AG)10 | TGGTGGTTGTTATGGGAGAAG | ATTTGTCTCTGTCTATTCCTTCCAC | 221 | [ 19] |
| BM140 | b04 | (GA)30 | TGCACAACACACATTTAGTGAC | CCTACCAAGATTGATTTATGGG | 190 | [ 19] |
| BMb43 | b04 | (TA)10 | GTGATCGGCTACATTAGCAT | GCTCTCATGTTCTCTTTCTCA | 143 | [ 20] |
| BM175 | b05 | (AT)5-(GA)19 | CAACAGTTAAAGGTCGTCAAATT | CCACTCTTAGCATCAACTGGA | 170 | [ 19] |
| BMb293 | b05 | (CTT)7 | CAATTCTACACTTTGGTGGG | AACGTCATTGATTTGACTCC | 154 | [ 20] |
| BM137 | b06 | (CT)33 | CGCTTACTCACTGTACGCACG | CCGTATCCGAGCACCGTAAC | 155 | [ 19] |
| BM170 | b06 | (CT)5-(CT)12 | AGCCAGGTGCAAGACCTTAG | AGATAGGGAGCTGGTGGTAGC | 179 | [ 18] |
| BMb526 | b07 | (TA)15 | AAAGGGCAAGTTAGATGTGA | TTTGAAGAATAGAAATCATACTG | 220 | [ 20] |
| BM201 | b07 | (GA)15 | TGGTGCTACAGACTTGATGG | TGTCACCTCTCTCCTCCAAT | 102 | [ 19] |
| BM189 | b08 | (CT)13 | CTCCCACTCTCACCCTCACT | GCGCCAAGTGAAACTAAGTAGA | 114 | [ 19] |
| BMd-44 | b08 | (AG)5 | GGCAGCTTACTAACCCGAAA | TTCCTTCCCCTTTCTTCTCC | 135 | [ 21] |
| BM141 | b09 | (GA)29 | TGAGGAGGAACAATGGTGGC | CTCACAAACCACAACGCACC | 218 | [ 19] |
| BM114 | b09 | (TA)8(GT)10 | AGCCTGGTGAAATGCTCATAG | CATGCTTGTTGCCTAACTCTCT | 234 | [ 18] |
| BM157 | b10 | (GA)16 | ACTTAACAAGGAATAGCCACACA | GTTAATTGTTTCCAATATCAACCTG | 113 | [ 18] |
| BMb356 | b01 | (TA)14 | TCCGAATTTCTTAATTTCACTT | ATCGCGGATTTATATGTGTC | 187 | [ 20] |
| BMb221 | b10 | (AT)10 | TGAAAGACAAGAGGGTTCAT | TTGTAGGCACTATTCCGTTT | 223 | [ 20] |
| BMd-41 | b11 | (ATT)9 | CAGTAAATATTGGCGTGGATGA | TGAAAGTGCAGAGTGGTGGA | 250 | [ 21] |
| BMb619 | b11 | (AT)22 | GATGGACACACTCACAAACA | TGTGTTCTACCACCAACAGA | 298 | [ 20] |
| GATS91 | b02 | (GA)11 | GAGTGCGGAAGCGAGTAGAG | TCCGTGTTCCTCTGTCTGTG | 229 | [ 19] |
| BM156 | b02 | (CT)32 | CTTGTTCCACCTCCCATCATAGC | TGCTTGCATCTCAGCCAGAATC | 267 | [ 19] |
| AG01 | b03 | (GA)8-(GA)5-(AG)4 | CATGCAGAGGAAGCAGAGTG | GAGCGTCGTCGTTTCGAT | 132 | [ 19] |
| BM172 | b03 | (GA)23 | CTGTAGCTCAAACAGGGCACT | GCAATACCGCCATGAGAGAT | 107 | [ 19] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klaedtke, S.M.; Caproni, L.; Klauck, J.; De la Grandville, P.; Dutartre, M.; Stassart, P.M.; Chable, V.; Negri, V.; Raggi, L. Short-Term Local Adaptation of Historical Common Bean (Phaseolus vulgaris L.) Varieties and Implications for In Situ Management of Bean Diversity. Int. J. Mol. Sci. 2017, 18, 493. https://doi.org/10.3390/ijms18030493
Klaedtke SM, Caproni L, Klauck J, De la Grandville P, Dutartre M, Stassart PM, Chable V, Negri V, Raggi L. Short-Term Local Adaptation of Historical Common Bean (Phaseolus vulgaris L.) Varieties and Implications for In Situ Management of Bean Diversity. International Journal of Molecular Sciences. 2017; 18(3):493. https://doi.org/10.3390/ijms18030493
Chicago/Turabian StyleKlaedtke, Stephanie M., Leonardo Caproni, Julia Klauck, Paul De la Grandville, Martin Dutartre, Pierre M. Stassart, Véronique Chable, Valeria Negri, and Lorenzo Raggi. 2017. "Short-Term Local Adaptation of Historical Common Bean (Phaseolus vulgaris L.) Varieties and Implications for In Situ Management of Bean Diversity" International Journal of Molecular Sciences 18, no. 3: 493. https://doi.org/10.3390/ijms18030493
APA StyleKlaedtke, S. M., Caproni, L., Klauck, J., De la Grandville, P., Dutartre, M., Stassart, P. M., Chable, V., Negri, V., & Raggi, L. (2017). Short-Term Local Adaptation of Historical Common Bean (Phaseolus vulgaris L.) Varieties and Implications for In Situ Management of Bean Diversity. International Journal of Molecular Sciences, 18(3), 493. https://doi.org/10.3390/ijms18030493







