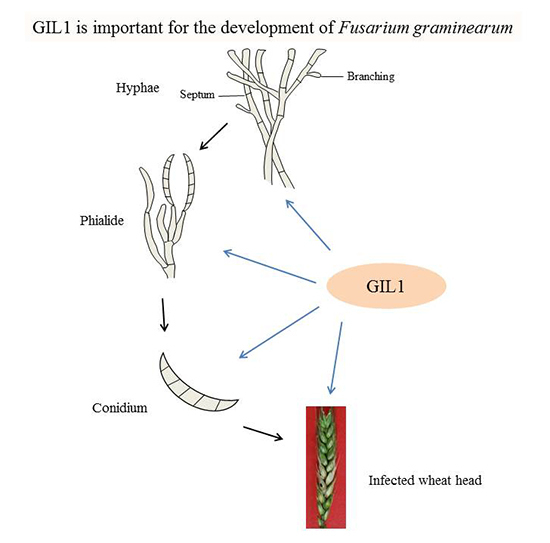
A Gin4-Like Protein Kinase GIL1 Involvement in Hyphal Growth, Asexual Development, and Pathogenesis in Fusarium graminearum
Abstract
:1. Introduction
2. Results
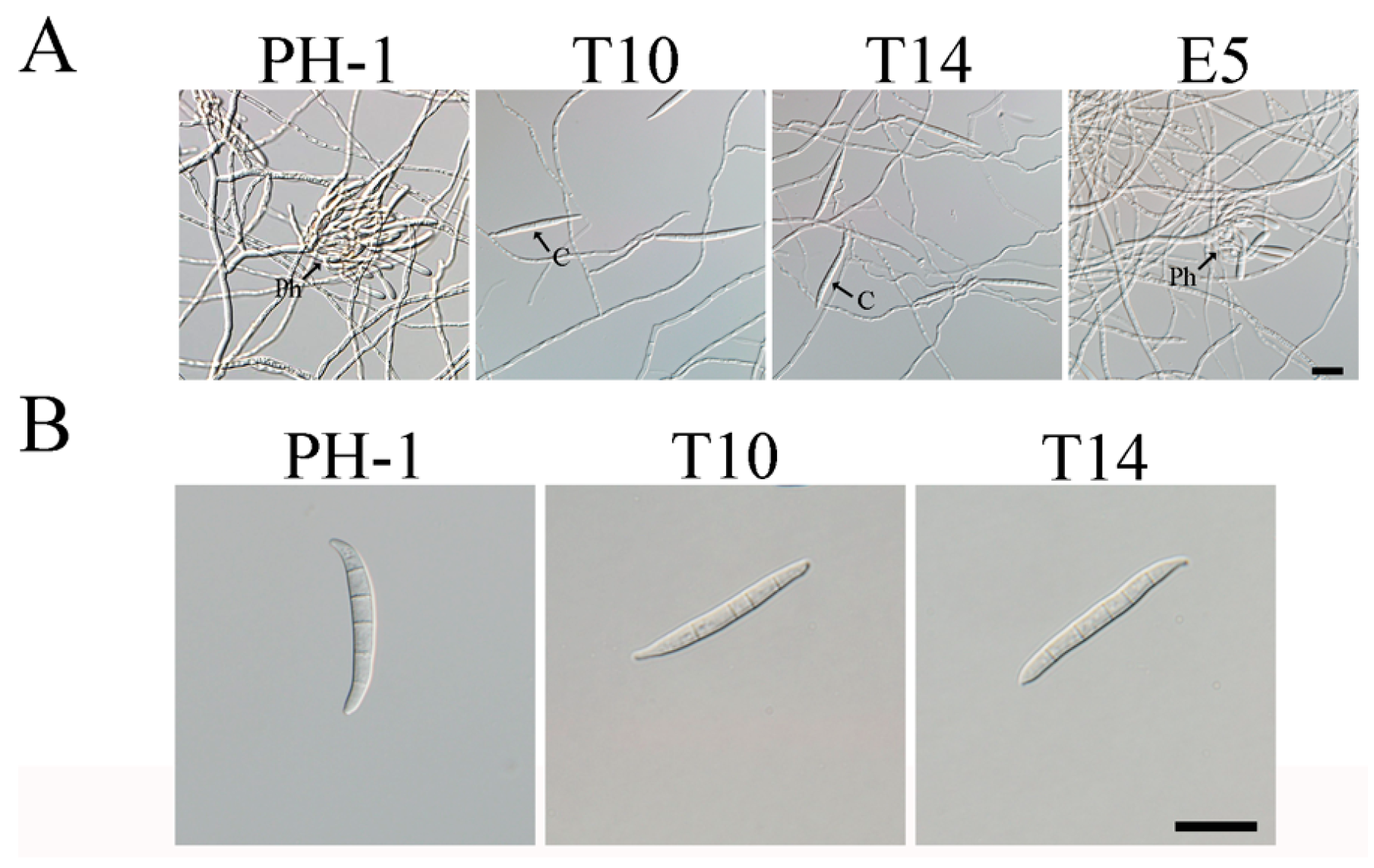
2.1. GIL1 Is Important for Normal Hyphal Growth
2.2. The Δgil1 Mutants Produce Straight, Longer Conidia Lacking Typical Foot Cells
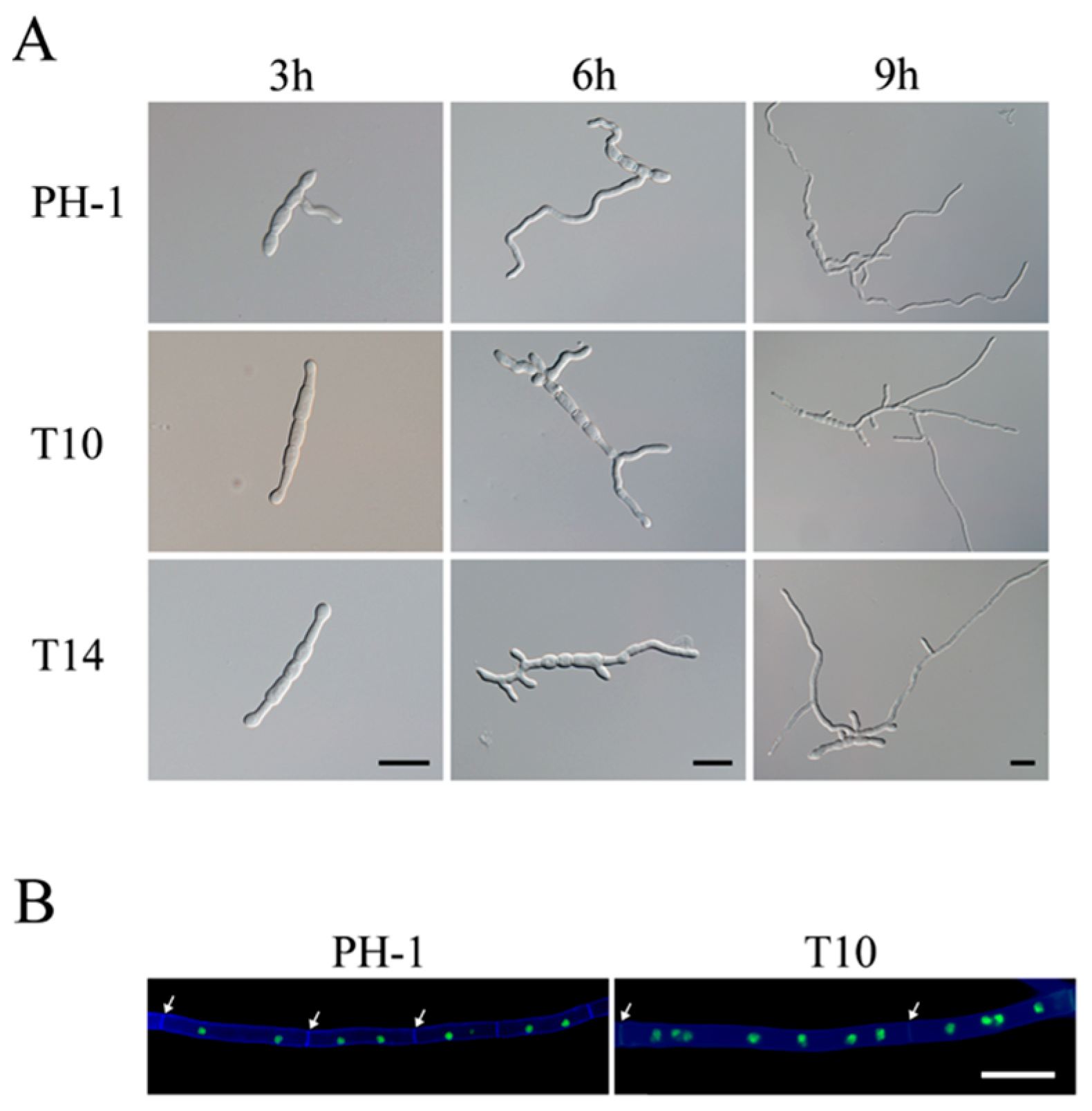
2.3. The Δgil1 Mutants Are Delayed in Conidium Germination but Increased in Germ Tube Branching
2.4. GIL1 Is Involved in Cytokinesis in Vegetative Hyphae
2.5. The Δgil1 Mutants Are Defective in Responses to Various Environmental Stresses
2.6. GIL1 Plays a Critical Role in Plant Infection in F. graminearum
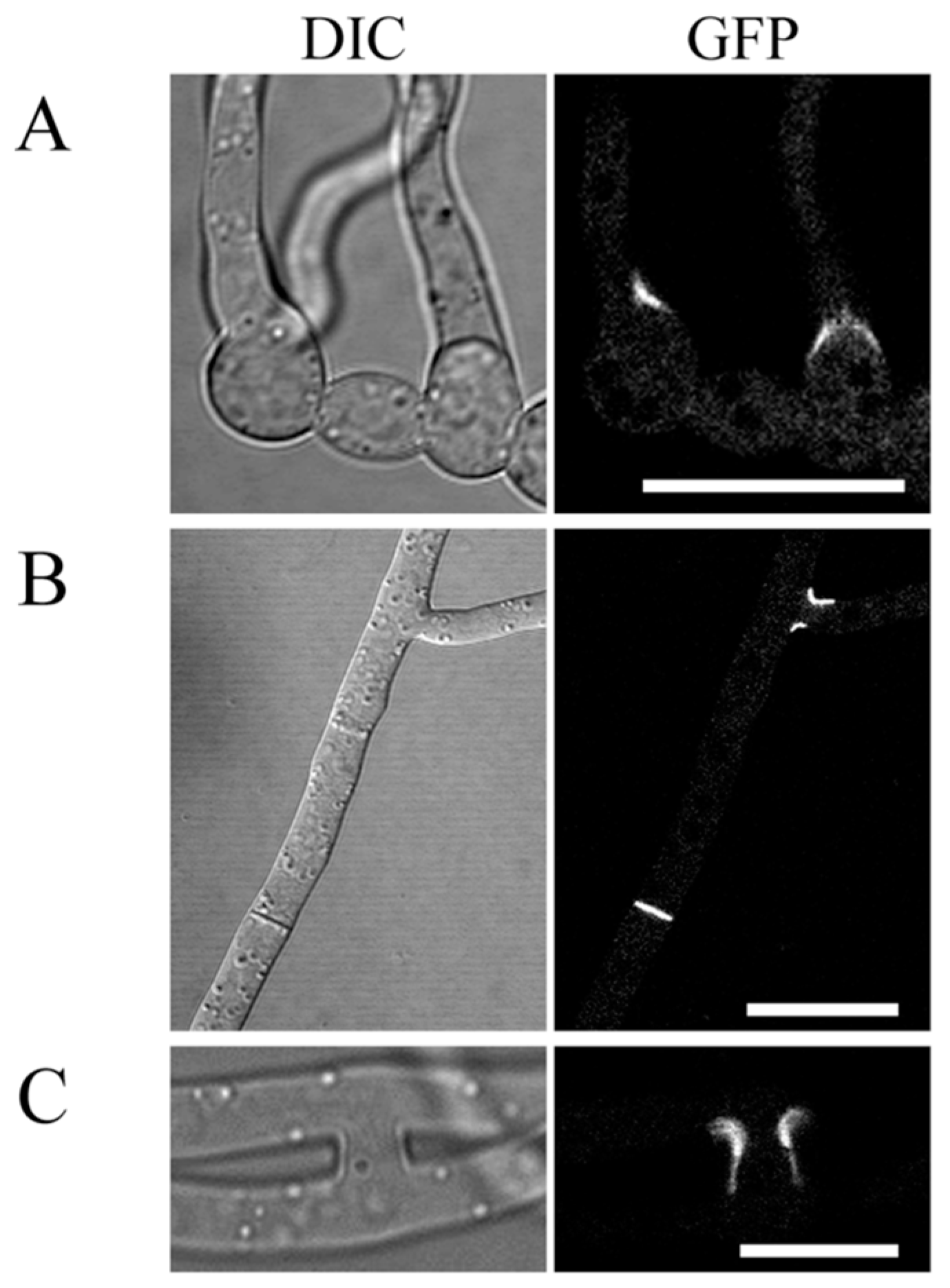
2.7. Localization of GIL1-GFP Fusion Proteins to Branching Point and Septa
3. Discussion
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. Identification of Δgil1 Mutants
4.3. Assays for Growth and Conidiation Defects
4.4. Plant Infection Assays
4.5. Generation of the GIL1-GFP Fusion Constructs
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dean, R.; van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, A.E. Gibberella from A (venaceae) to Z (eae). Annu. Rev. Phytopathol. 2003, 41, 177–198. [Google Scholar] [CrossRef] [PubMed]
- De Walle, J.V.; Sergent, T.; Piront, N.; Toussaint, O.; Schneider, Y.J.; Larondelle, Y. Deoxynivalenol affects in vitro intestinal epithelial cell barrier integrity through inhibition of protein synthesis. Toxicol. Appl. Pharmacol. 2010, 245, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.H.; Desjardins, A.E.; Plattner, R.D. Deoxynivalenol-nonproducing Fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia 2002, 153, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.H.; Hohn, T.M.; McCormick, S.P. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol. Plant Microbe Interact. 1995, 8, 593–601. [Google Scholar] [PubMed]
- Desjardins, A.E.; Bai, G.; Plattner, R.D.; Proctor, R.H. Analysis of aberrant virulence of Gibberella zeae following transformation-mediated complementation of a trichothecene-deficient (Tri5) mutant. Microbiology 2000, 146, 2059–2068. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wang, Y.; Zhou, X.; Wang, Y.; Xu, J.R. The Sch9 kinase regulates conidium size, stress responses, and pathogenesis in Fusarium graminearum. PLoS ONE 2014, 9, e105811. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Zhou, X.; Gu, X.; Cao, S.; Wang, C.; Xu, J.R. The cAMP-PKA pathway regulates growth, sexual and asexual differentiation, and pathogenesis in Fusarium graminearum. Mol. Plant Microbe Interact. 2014, 27, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Zhang, S.; Zhang, Q.; Tao, Y.; Wang, C.; Xu, J.R. FgSKN7 and FgATF1 have overlapping functions in ascosporogenesis, pathogenesis and stress responses in Fusarium graminearum. Environ. Microbiol. 2015, 17, 1245–1260. [Google Scholar] [CrossRef] [PubMed]
- Son, H.; Seo, Y.S.; Min, K.; Park, A.R.; Lee, J.; Jin, J.M.; Lin, Y.; Cao, P.; Hong, S.Y.; Kim, E.K.; et al. A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum. PLoS Pathog. 2011, 7, e1002310. [Google Scholar] [CrossRef] [PubMed]
- Min, K.; Shin, Y.; Son, H.; Lee, J.; Kim, J.C.; Choi, G.J.; Lee, Y.W. Functional analyses of the nitrogen regulatory gene areA in Gibberella zeae. FEMS Microbiol. Lett. 2012, 334, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, W.; Hou, Z.; Wang, C.; Zhou, X.; Jonkers, W.; Ding, S.; Kistler, H.C.; Xu, J.R. A novel transcriptional factor important for pathogenesis and ascosporogenesis in Fusarium graminearum. Mol. Plant Microbe Interact. 2011, 24, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, S.; Hou, R.; Zhao, Z.; Zheng, Q.; Xu, Q.; Zheng, D.; Wang, G.; Liu, H.; Gao, X.; et al. Functional analysis of the kinome of the wheat scab fungus Fusarium graminearum. PLoS Pathog. 2011, 7, e1002460. [Google Scholar] [CrossRef] [PubMed]
- Seong, K.Y.; Pasquali, M.; Zhou, X.; Song, J.; Hilburn, K.; McCormick, S.; Dong, Y.; Xu, J.R.; Kistler, H.C. Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Mol. Microbiol. 2009, 72, 354–367. [Google Scholar] [CrossRef] [PubMed]
- Blumke, A.; Falter, C.; Herrfurth, C.; Sode, B.; Bode, R.; Schafer, W.; Feussner, I.; Voigt, C.A. Secreted fungal effector lipase releases free fatty acids to inhibit innate immunity-related callose formation during wheat head infection. Plant Physiol. 2014, 165, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Voigt, C.A.; Schafer, W.; Salomon, S. A secreted lipase of Fusarium graminearum is a virulence factor required for infection of cereals. Plant J. 2005, 42, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Seong, K.; Hou, Z.; Tracy, M.; Kistler, H.C.; Xu, J.R. Random insertional mutagenesis identifies genes associated with virulence in the wheat scab fungus Fusarium graminearum. Phytopathology 2005, 95, 744–750. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, J.; Xu, J.; Shi, J. Fgilv5 is required for branched-chain amino acid biosynthesis and full virulence in Fusarium graminearum. Microbiology 2014, 160, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.; Mott, E.; Farley, T.; Hammond-Kosack, K. The Fusarium graminearum MAP1 gene is essential for pathogenicity and development of perithecia. Mol. Plant Pathol. 2003, 4, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Jenczmionka, N.J.; Maier, F.J.; Losch, A.P.; Schafer, W. Mating, conidiation and pathogenicity of Fusarium graminearum, the main causal agent of the head-blight disease of wheat, are regulated by the MAP kinase GPMK1. Curr. Genet. 2003, 43, 87–95. [Google Scholar] [PubMed]
- Zheng, D.; Zhang, S.; Zhou, X.; Wang, C.; Xiang, P.; Zheng, Q.; Xu, J.R. The FgHOG1 pathway regulates hyphal growth, stress responses, and plant infection in Fusarium graminearum. PLoS ONE 2012, 7, e49495. [Google Scholar] [CrossRef] [PubMed]
- Longtine, M.S.; Fares, H.; Pringle, J.R. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J. Cell Biol. 1998, 143, 719–736. [Google Scholar] [CrossRef] [PubMed]
- McMillan, J.N.; Longtine, M.S.; Sia, R.A.; Theesfeld, C.L.; Bardes, E.S.; Pringle, J.R.; Lew, D.J. The morphogenesis checkpoint in Saccharomyces cerevisiae: Cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol. Cell. Biol. 1999, 19, 6929–6939. [Google Scholar] [CrossRef] [PubMed]
- Longtine, M.S.; Theesfeld, C.L.; McMillan, J.N.; Weaver, E.; Pringle, J.R.; Lew, D.J. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000, 20, 4049–4061. [Google Scholar] [CrossRef] [PubMed]
- Altman, R.; Kellogg, D. Control of mitotic events by Nap1 and the Gin4 kinase. J. Cell Biol. 1997, 138, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Okuzaki, D.; Watanabe, T.; Tanaka, S.; Nojima, H. The Saccharomyces cerevisiae bud-neck proteins Kcc4 and Gin4 have distinct but partially-overlapping cellular functions. Genes Genet. Syst. 2003, 78, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Morrell, J.L.; Nichols, C.B.; Gould, K.L. The GIN4 family kinase, Cdr2p, acts independently of septins in fission yeast. J. Cell Sci. 2004, 117, 5293–5302. [Google Scholar] [CrossRef] [PubMed]
- Breeding, C.S.; Hudson, J.; Balasubramanian, M.K.; Hemmingsen, S.M.; Young, P.G.; Gould, K.L. The cdr2+ gene encodes a regulator of G2/M progression and cytokinesis in Schizosaccharomyces pombe. Mol. Biol. Cell 1998, 9, 3399–3415. [Google Scholar] [CrossRef] [PubMed]
- Kanoh, J.; Russell, P. The protein kinase Cdr2, related to Nim1/Cdr1 mitotic inducer, regulates the onset of mitosis in fission yeast. Mol. Biol. Cell 1998, 9, 3321–3334. [Google Scholar] [CrossRef]
- Wightman, R.; Bates, S.; Amornrrattanapan, P.; Sudbery, P. In Candida albicans, the Nim1 kinases Gin4 and hsl1 negatively regulate pseudohypha formation and Gin4 also controls septin organization. J. Cell Biol. 2004, 164, 581–591. [Google Scholar] [CrossRef] [PubMed]
- DeMay, B.S.; Meseroll, R.A.; Occhipinti, P.; Gladfelter, A.S. Regulation of distinct septin rings in a single cell by Elm1p and Gin4p kinases. Mol. Biol. Cell 2009, 20, 2311–2326. [Google Scholar] [CrossRef] [PubMed]
- De Souza, C.P.; Hashmi, S.B.; Osmani, A.H.; Andrews, P.; Ringelberg, C.S.; Dunlap, J.C.; Osmani, S.A. Functional analysis of the Aspergillus nidulans kinome. PLoS ONE 2013, 8, e58008. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.J.; Lu, Q.; Grunstein, M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 1996, 10, 1327–1340. [Google Scholar] [CrossRef] [PubMed]
- Barral, Y.; Parra, M.; Bidlingmaier, S.; Snyder, M. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 1999, 13, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zhang, H.; Qi, L.; Zhang, S.; Zhou, X.; Zhang, Y.; Xu, J.R. Fgkin1 kinase localizes to the septal pore and plays a role in hyphal growth, ascospore germination, pathogenesis, and localization of Tub1 beta-tubulins in Fusarium graminearum. New Phytol. 2014, 204, 943–954. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lee, J.; Lee, S.; Park, E.H.; Kim, K.W.; Kim, M.D.; Yun, S.H.; Lee, Y.W. Gzsnf1 is required for normal sexual and asexual development in the ascomycete Gibberella zeae. Eukaryot. Cell 2009, 8, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, R.; Cowden, S.; Hernandez-Rodriguez, Y.; Momany, M. Septins AspA and AspC are important for normal development and limit the emergence of new growth foci in the multicellular fungus Aspergillus nidulans. Eukaryot. Cell 2010, 9, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Boenisch, M.J.; Schafer, W. Fusarium graminearum forms mycotoxin producing infection structures on wheat. BMC Plant Biol. 2011, 11, 110. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.X.; Zhao, P.; Zeng, G.S.; Wang, Y.M.; Sudbery, I.; Wang, Y. Phosphoregulation of Nap1 plays a role in septin ring dynamics and morphogenesis in Candida albicans. MBio 2014, 5, e00915-13. [Google Scholar] [CrossRef] [PubMed]
- Okuzaki, D.; Tanaka, S.; Kanazawa, H.; Nojima, H. Gin4 of S. cerevisiae is a bud neck protein that interacts with the Cdc28 complex. Genes Cells 1997, 2, 753–770. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Jacobson, D.J.; Shiu, P.K. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annu. Rev. Genet. 2000, 34, 165–186. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Xue, C.; Peng, Y.; Katan, T.; Kistler, H.C.; Xu, J.R. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant Microbe Interact. 2002, 15, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Seong, K.; Li, L.; Hou, Z.; Tracy, M.; Kistler, H.C.; Xu, J.R. Cryptic promoter activity in the coding region of the HMG-CoA reductase gene in Fusarium graminearum. Fungal Genet. Biol. 2006, 43, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Heyer, C.; Choi, Y.E.; Mehrabi, R.; Xu, J.R. The CID1 cyclin C-like gene is important for plant infection in Fusarium graminearum. Fungal Genet. Biol. 2010, 47, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Gale, L.R.; Chen, L.F.; Hernick, C.A.; Takamura, K.; Kistler, H.C. Population analysis of Fusarium graminearum from wheat fields in eastern China. Phytopathology 2002, 92, 1315–1322. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.E.; Xu, J.R. The camp signaling pathway in Fusarium verticillioides is important for conidiation, plant infection, and stress responses but not fumonisin production. Mol. Plant Microbe Interact. 2010, 23, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, G.; Xu, J.R. Efficient approaches for generating GFP fusion and epitope-tagging constructs in filamentous fungi. Methods Mol. Biol. 2011, 722, 199–212. [Google Scholar] [PubMed]
- Bruno, K.S.; Tenjo, F.; Li, L.; Hamer, J.E.; Xu, J.R. Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot. Cell 2004, 3, 1525–1532. [Google Scholar] [CrossRef] [PubMed]
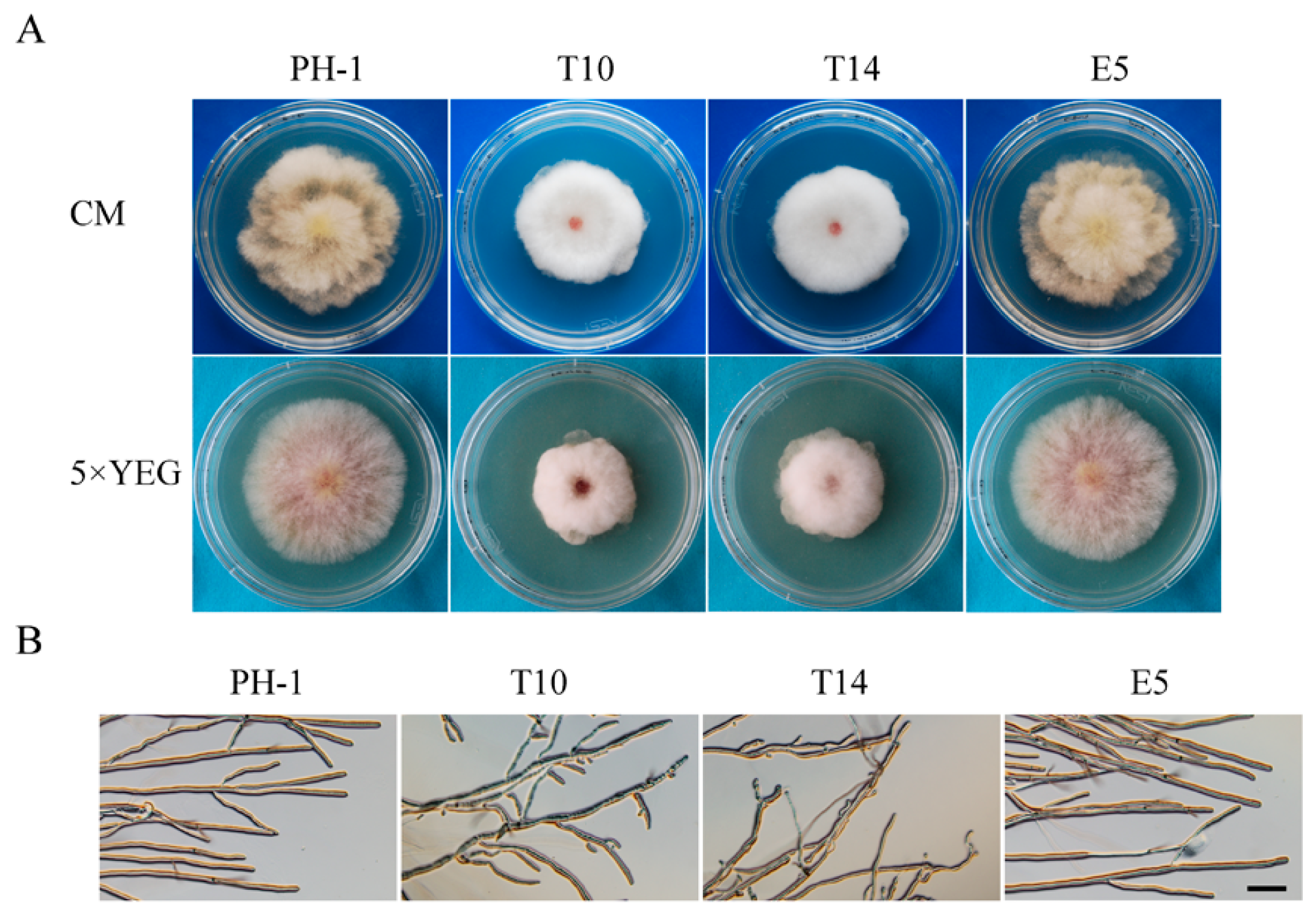

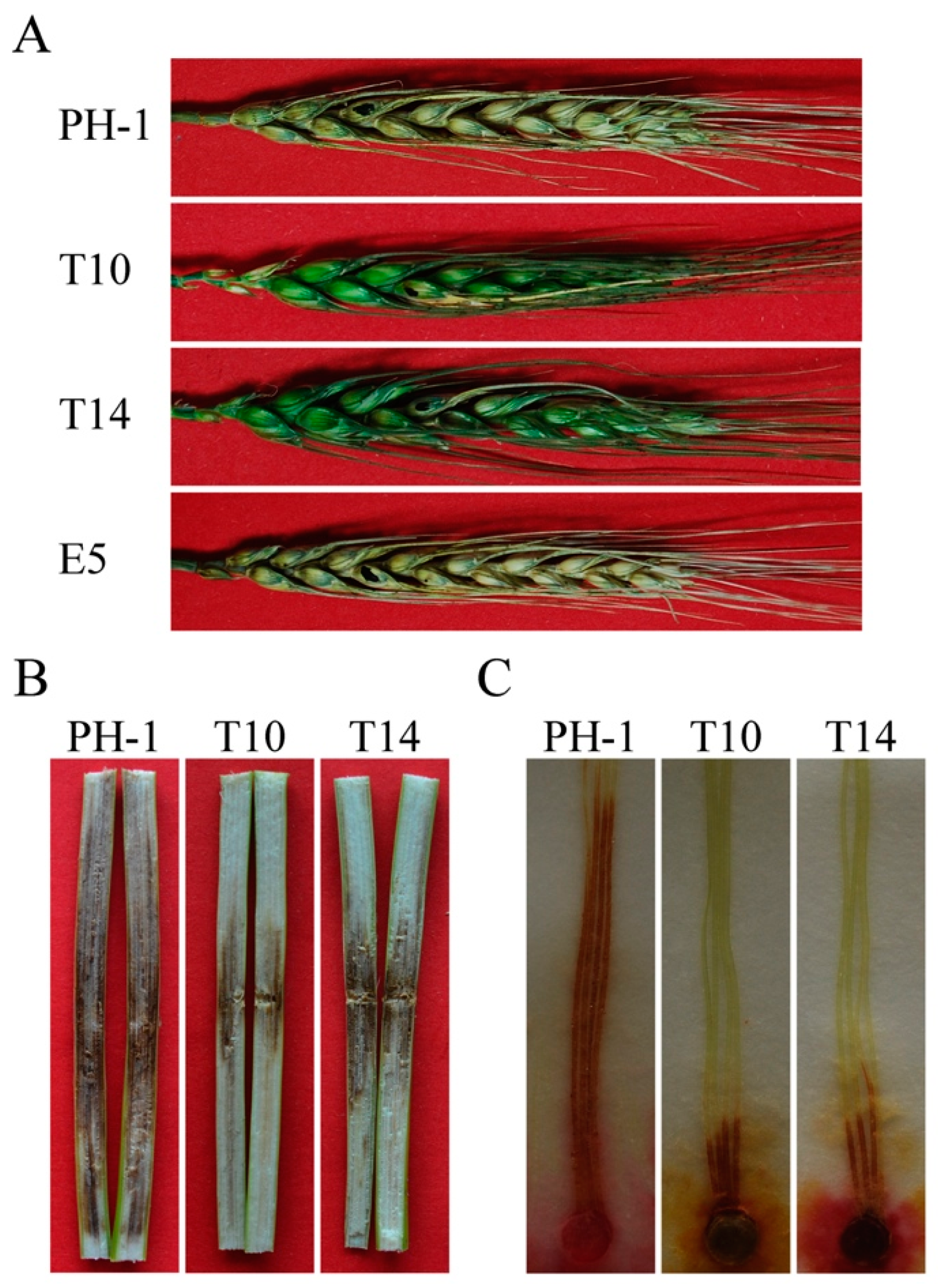
| Strain | Growth Rate (cm/day) a | Conidiation (106 conidia/mL) | Conidium Size (µm) | |||
|---|---|---|---|---|---|---|
| PDA | CM | 5× YEG | Length | Width | ||
| PH-1 | 1.3 ± 0.0 A | 1.0 ± 0.0 A | 1.0 ± 0.0 A | 1.2 ± 0.1A | 41.6 ± 0.7 B | 5.4 ± 0.2 A |
| T10 | 0.8 ± 0.0 B | 0.8 ± 0.0 B | 0.7 ± 0.0 B | 0.7 ± 0.1B | 49.2 ± 2.8 A | 5.4 ± 0.2 A |
| T14 | 0.8 ± 0.0 B | 0.8 ± 0.0 B | 0.7 ± 0.0 B | 0.8 ± 0.1B | 47.5 ± 2.0 A | 5.3 ± 0.4 A |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, D.; Zhang, S.; Li, X.; Xu, J.-R.; Schultzhaus, Z.; Jin, Q. A Gin4-Like Protein Kinase GIL1 Involvement in Hyphal Growth, Asexual Development, and Pathogenesis in Fusarium graminearum. Int. J. Mol. Sci. 2017, 18, 424. https://doi.org/10.3390/ijms18020424
Yu D, Zhang S, Li X, Xu J-R, Schultzhaus Z, Jin Q. A Gin4-Like Protein Kinase GIL1 Involvement in Hyphal Growth, Asexual Development, and Pathogenesis in Fusarium graminearum. International Journal of Molecular Sciences. 2017; 18(2):424. https://doi.org/10.3390/ijms18020424
Chicago/Turabian StyleYu, Dan, Shijie Zhang, Xiaoping Li, Jin-Rong Xu, Zachary Schultzhaus, and Qiaojun Jin. 2017. "A Gin4-Like Protein Kinase GIL1 Involvement in Hyphal Growth, Asexual Development, and Pathogenesis in Fusarium graminearum" International Journal of Molecular Sciences 18, no. 2: 424. https://doi.org/10.3390/ijms18020424