Significant Down-Regulation of “Biological Adhesion” Genes in Porcine Oocytes after IVM
Abstract
:1. Introduction
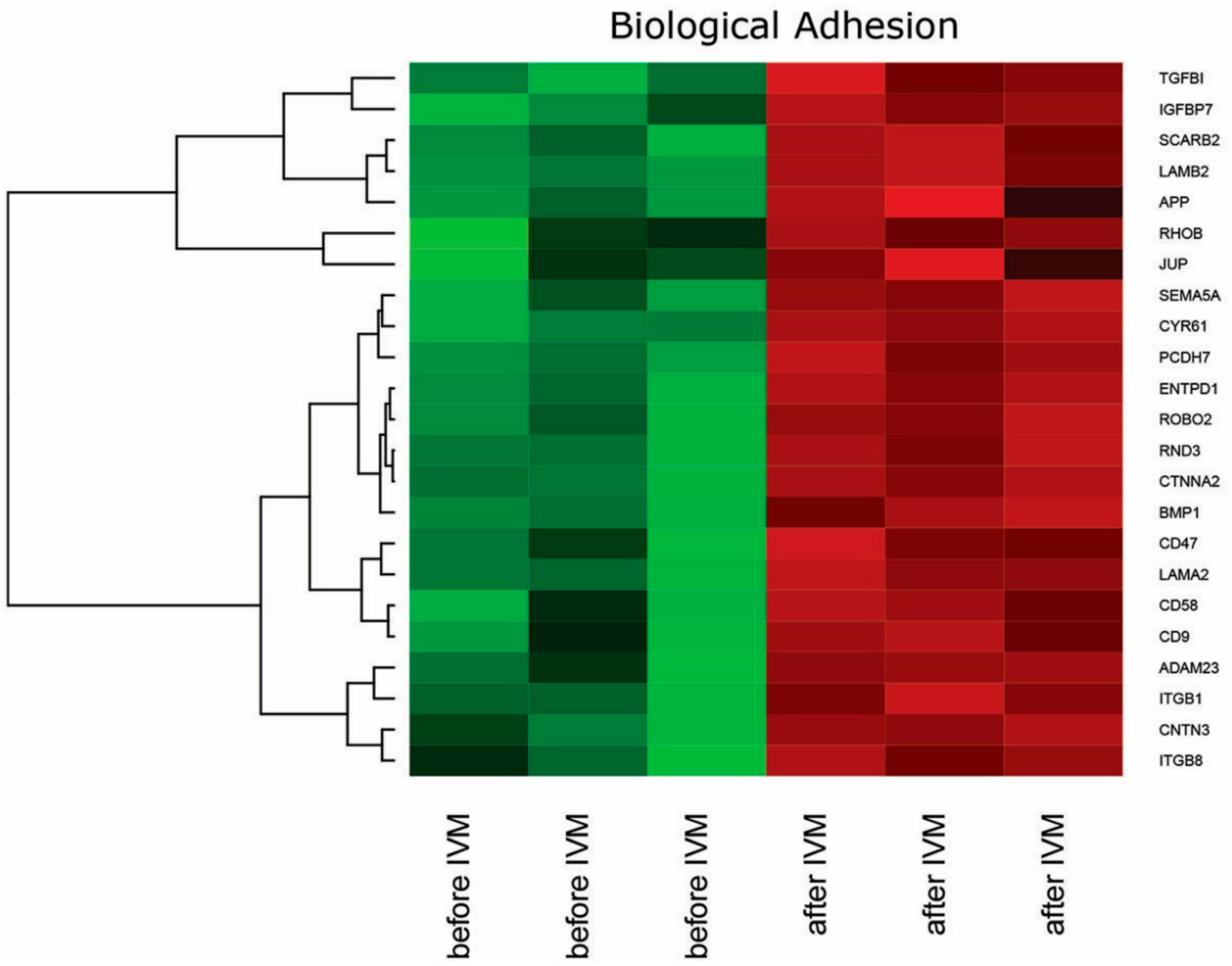
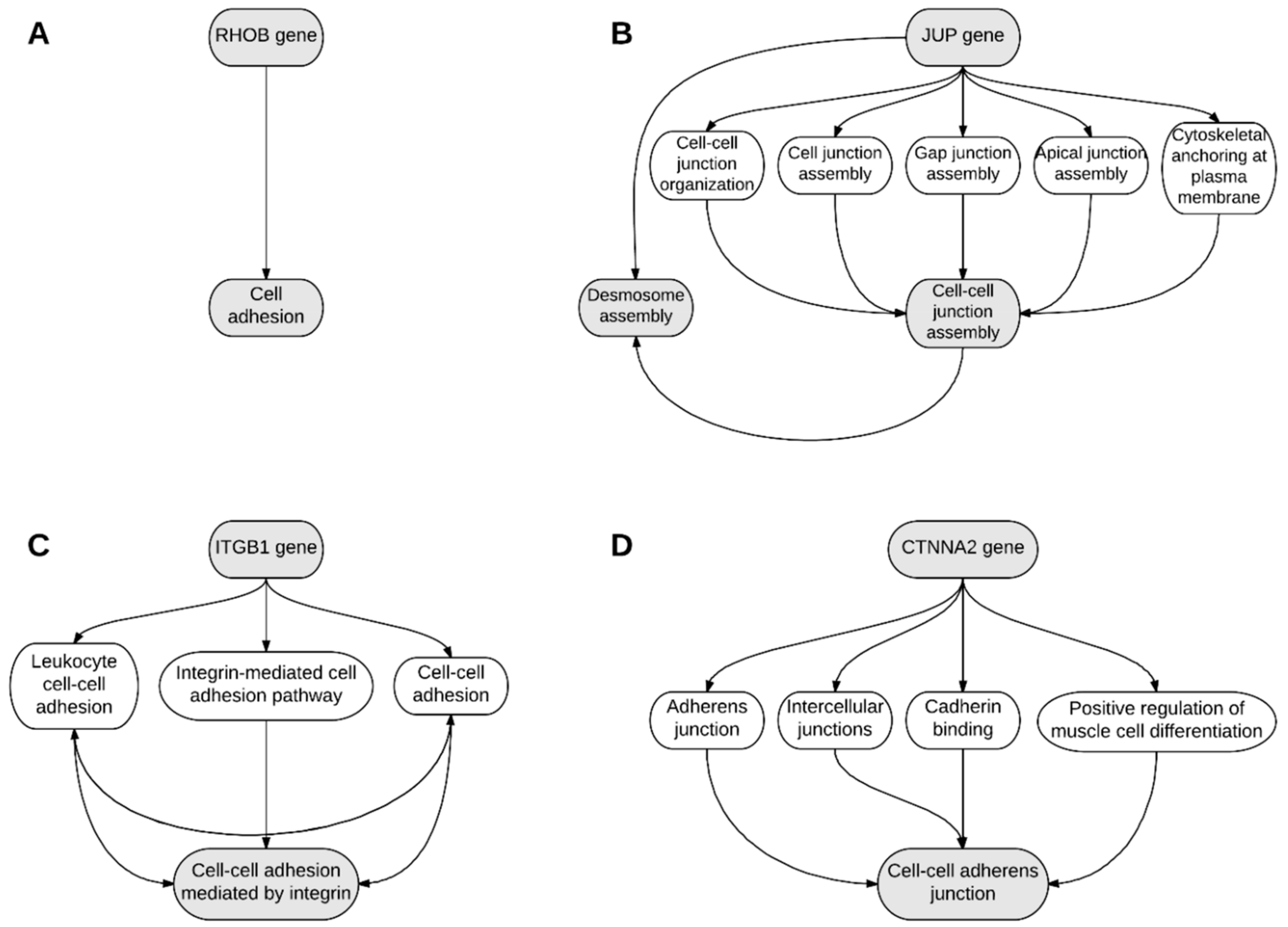
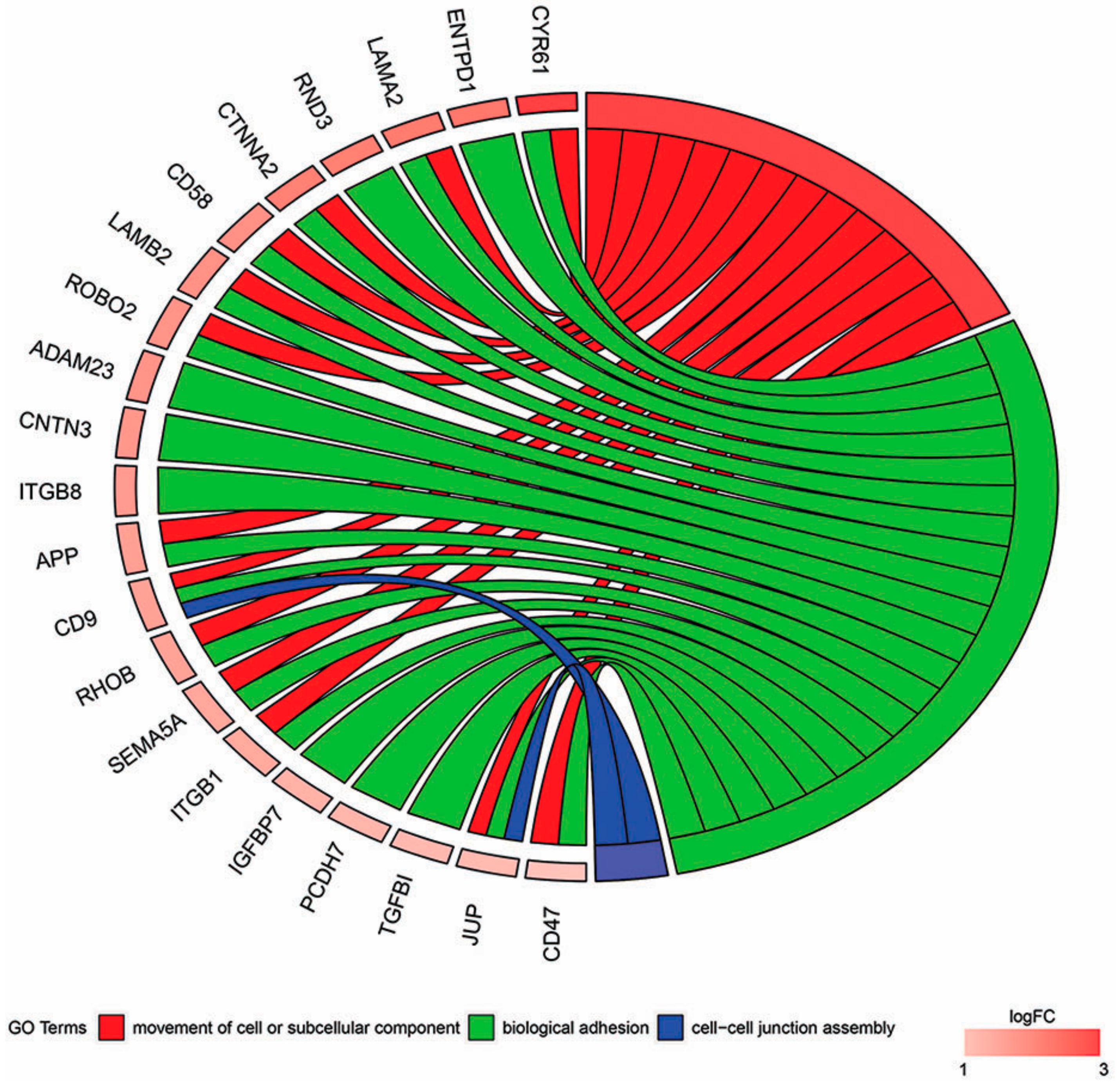
2. Results
3. Discussion
4. Materials and Methods
4.1. Experimental Design
4.2. Animals
4.3. Collection of Porcine Ovaries and Cumulus–Oocyte Complexes (COCs)
4.4. Assessment of Oocyte Developmental Competence by BCB Test
4.5. In Vitro Maturation of Porcine Cumulus-Oocyte-Complexes (COCs)
4.6. RNA Extraction from Porcine Oocytes
4.7. Microarray Expression Analysis and Statistics
4.8. Validation of Microarray Results with Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Uyar, A.; Torrealday, S.; Seli, E. Cumulus and granulosa cell markers of oocyte and embryo quality. Fertil. Steril. 2013, 99, 979–997. [Google Scholar] [CrossRef] [PubMed]
- Orisaka, M.; Tajima, K.; Tsang, B.K.; Kotsuji, F. Oocyte-granulosa-theca cell interactions during preantral follicular development. J. Ovarian Res. 2009, 2. [Google Scholar] [CrossRef] [PubMed]
- Eppig, J.J. Coordination of nuclear and cytoplasmic oocyte maturation in eutherian mammals. Reprod. Fertil. Dev. 1996, 8, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Kranc, W.; Celichowski, P.; Budna, J.; Khozmi, R.; Bryja, A.; Ciesiółka, S.; Rybska, M.; Borys, S.; Jeseta, M.; Bukowska, D.; et al. Positive regulation of macromolecule metabolic process belongs to the main mechanisms crucial for porcine oocytes maturation. Adv. Cell Biol. 2017, 5, 15–32. [Google Scholar] [CrossRef]
- Jin, J.X.; Lee, S.; Khoirinaya, C.; Oh, A.; Kim, G.A.; Lee, B.C. Supplementation with spermine during in vitro maturation of porcine oocytes improves early embryonic development after parthenogenetic activation and somatic cell nuclear transfer. J. Anim. Sci. 2016, 94, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Galeati, G.; Giaretta, E.; Zannoni, A.; Bucci, D.; Tamanini, C.; Forni, M.; Spinaci, M. Embelin supplementation of in vitro maturation medium does not influence nuclear and cytoplasmic maturation of pig oocytes. J. Physiol. Pharmacol. 2016, 67, 513–519. [Google Scholar] [PubMed]
- Wu, J.; Emery, B.R.; Carrell, D.T. In vitro growth, maturation, fertilization, and embryonic development of oocytes from porcine preantral follicles. Biol. Reprod. 2001, 64, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Budna, J.; Bryja, A.; Celichowski, P.; Kranc, W.; Ciesiolka, S.; Borys, S.; Rybska, M.; Kolecka-Bednarczyk, A.; Jeseta, M.; Bukowska, D.; et al. “Bone Development” Is an Ontology Group Upregulated in Porcine Oocytes before In Vitro Maturation: A Microarray Approach. DNA Cell Biol. 2017, 36, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Budna, J.; Bryja, A.; Celichowski, P.; Kahan, R.; Kranc, W.; Ciesiolka, S.; Rybska, M.; Borys, S.; Jeseta, M.; Bukowska, D.; et al. Genes of cellular components of morphogenesis in porcine oocytes before and after IVM. Reproduction 2017, 154, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Budna, J.; Rybska, M.; Ciesiolka, S.; Bryja, A.; Borys, S.; Kranc, W.; Wojtanowicz-Markiewicz, K.; Jeseta, M.; Sumelka, E.; Bukowska, D.; et al. Expression of genes associated with BMP signaling pathway in porcine oocytes before and after IVM—A microarray approach. Reprod. Biol. Endocrinol. 2017, 15. [Google Scholar] [CrossRef] [PubMed]
- Kranc, W.; Budna, J.; Chachula, A.; Borys, S.; Bryja, A.; Rybska, M.; Ciesiolka, S.; Sumelka, E.; Jeseta, M.; Brussow, K.P.; et al. “Cell Migration” Is the Ontology Group Differentially Expressed in Porcine Oocytes Before and After In Vitro Maturation: A Microarray Approach. DNA Cell Biol. 2017, 36, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Budna, J.; Celichowski, P.; Karimi, P.; Kranc, W.; Bryja, A.; Ciesiółka, S.; Rybska, M.; Borys, S.; Jeseta, M.; Bukowska, D.; et al. Does Porcine oocytes maturation in vitro is regulated by genes involved in transforming growth factor beta receptor signaling pathway? Adv. Cell Biol. 2017, 5, 1–14. [Google Scholar] [CrossRef]
- Cal, S.; Freije, J.M.; Lopez, J.M.; Takada, Y.; Lopez-Otin, C. ADAM 23/MDC3, a human disintegrin that promotes cell adhesion via interaction with the alphavbeta3 integrin through an RGD-independent mechanism. Mol. Biol. Cell 2000, 11, 1457–1469. [Google Scholar] [CrossRef] [PubMed]
- Tsanou, E.; Peschos, D.; Batistatou, A.; Charalabopoulos, A.; Charalabopoulos, K. The E-cadherin adhesion molecule and colorectal cancer. A global literature approach. Anticancer Res. 2008, 28, 3815–3826. [Google Scholar] [PubMed]
- Wang, X.; Tully, O.; Ngo, B.; Zitin, M.; Mullin, J.M. Epithelial tight junctional changes in colorectal cancer tissues. Sci. World J. 2011, 11, 826–841. [Google Scholar] [CrossRef] [PubMed]
- Turksen, K.; Troy, T.-C. Junctions gone bad: Claudins and loss of the barrier in cancer. Biochim. Biophys. Acta 2011, 1816, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Kanczuga-Koda, L. Gap junctions and their role in physiology and pathology of the digestive tract. Postepy Hig. Med. Dosw. 2004, 58, 158–165. [Google Scholar]
- Kempisty, B.; Ziolkowska, A.; Ciesiolka, S.; Piotrowska, H.; Antosik, P.; Bukowska, D.; Nowicki, M.; Brussow, K.P.; Zabel, M. Study on connexin gene and protein expression and cellular distribution in relation to real-time proliferation of porcine granulosa cells. J. Biol. Regul. Homeost. Agents 2014, 28, 625–635. [Google Scholar] [PubMed]
- Tatone, C.; Carbone, M.C. Possible involvement of integrin-mediated signalling in oocyte activation: Evidence that a cyclic RGD-containing peptide can stimulate protein kinase C and cortical granule exocytosis in mouse oocytes. Reprod. Biol. Endocrinol. 2006, 4. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.P.; Schultz, R.M.; Kopf, G.S. Identification and localization of integrin subunits in oocytes and eggs of the mouse. Mol. Reprod. Dev. 1995, 40, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Krisher, R.L. In vitro maturation (IVM) of porcine oocytes. Methods Mol. Biol. 2012, 825, 183–198. [Google Scholar] [PubMed]
- Dunning, K.R.; Lane, M.; Brown, H.M.; Yeo, C.; Robker, R.L.; Russell, D.L. Altered composition of the cumulus-oocyte complex matrix during in vitro maturation of oocytes. Hum. Reprod. 2007, 22, 2842–2850. [Google Scholar] [CrossRef] [PubMed]
- Budna, J.; Chachula, A.; Kazmierczak, D.; Rybska, M.; Ciesiolka, S.; Bryja, A.; Kranc, W.; Borys, S.; Zok, A.; Bukowska, D.; et al. Morphogenesis-related gene-expression profile in porcine oocytes before and after in vitro maturation. Zygote 2017, 25, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Arias-Alvarez, M.; Garcia-Garcia, R.M.; Lopez-Tello, J.; Rebollar, P.G.; Gutierrez-Adan, A.; Lorenzo, P.L. In vivo and in vitro maturation of rabbit oocytes differently affects the gene expression profile, mitochondrial distribution, apoptosis and early embryo development. Reprod. Fertil. Dev. 2017, 29, 1667–1679. [Google Scholar] [CrossRef] [PubMed]
- Yerushalmi, G.M.; Maman, E.; Yung, Y.; Kedem, A.; Hourvitz, A. Molecular characterization of the human ovulatory cascade-lesson from the IVF/IVM model. J. Assist. Reprod. Genet. 2011, 28, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Hao, C.; Shen, X.; Zhang, Y.; Liu, X. RUNX2, GPX3 and PTX3 gene expression profiling in cumulus cells are reflective oocyte/embryo competence and potentially reliable predictors of embryo developmental competence in PCOS patients. Reprod. Biol. Endocrinol. 2013, 11. [Google Scholar] [CrossRef] [PubMed]
- Dieci, C.; Lodde, V.; Labreque, R.; Dufort, I.; Tessaro, I.; Sirard, M.-A.; Luciano, A.M. Differences in cumulus cell gene expression indicate the benefit of a pre-maturation step to improve in-vitro bovine embryo production. Mol. Hum. Reprod. 2016, 22, 882–897. [Google Scholar] [PubMed]
- Kempisty, B.; Ziolkowska, A.; Piotrowska, H.; Ciesiolka, S.; Antosik, P.; Bukowska, D.; Zawierucha, P.; Wozna, M.; Jaskowski, J.M.; Brussow, K.P.; et al. Short-term cultivation of porcine cumulus cells influences the cyclin-dependent kinase 4 (Cdk4) and connexin 43 (Cx43) protein expression—A real-time cell proliferation approach. J. Reprod. Dev. 2013, 59, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Kempisty, B.; Ziolkowska, A.; Piotrowska, H.; Zawierucha, P.; Antosik, P.; Bukowska, D.; Ciesiolka, S.; Jaskowski, J.M.; Brussow, K.P.; Nowicki, M.; et al. Real-time proliferation of porcine cumulus cells is related to the protein levels and cellular distribution of Cdk4 and Cx43. Theriogenology 2013, 80, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.; Albertini, D.F. Gap junctions between the oocyte and companion follicle cells in the mammalian ovary. J. Cell Biol. 1976, 71, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Guasch, R.M.; Scambler, P.; Jones, G.E.; Ridley, A.J. RhoE regulates actin cytoskeleton organization and cell migration. Mol. Cell Biol. 1998, 18, 4761–4771. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.; Brennan, M.; Moran, N. Integrins as therapeutic targets: Lessons and opportunities. Nat. Rev. Drug Discov. 2010, 9, 804–820. [Google Scholar] [CrossRef] [PubMed]
- Lau, L.F. CCN1/CYR61: The very model of a modern matricellular protein. Cell Mol. Life Sci. 2011, 68, 3149–3163. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-W.; Bae, J.-S.; Kim, K.-S.; Park, S.-H.; Lee, B.-H.; Choi, J.-Y.; Park, J.-Y.; Ha, S.-W.; Kim, Y.-L.; Kwon, T.-H.; et al. Beta ig-h3 promotes renal proximal tubular epithelial cell adhesion, migration and proliferation through the interaction with alpha3beta1 integrin. Exp. Mol. Med. 2004, 36, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Thumkeo, D.; Watanabe, S.; Narumiya, S. Physiological roles of Rho and Rho effectors in mammals. Eur. J. Cell Biol. 2013, 92, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Villalonga, P.; Guasch, R.M.; Riento, K.; Ridley, A.J. RhoE inhibits cell cycle progression and Ras-induced transformation. Mol. Cell Biol. 2004, 24, 7829–7840. [Google Scholar] [CrossRef] [PubMed]
- Nousbeck, J.; Sarig, O.; Avidan, N.; Indelman, M.; Bergman, R.; Ramon, M.; Enk, C.D.; Sprecher, E. Insulin-like growth factor-binding protein 7 regulates keratinocyte proliferation, differentiation and apoptosis. J. Investig. Dermatol. 2010, 130, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Jung, M.-Y.; Kim, I.-S. Stabilin-2 mediates homophilic cell–cell interactions via its FAS1 domains. FEBS Lett. 2009, 583, 1375–1380. [Google Scholar] [CrossRef] [PubMed]
- Zhurinsky, J.; Shtutman, M.; Ben-Ze’ev, A. Plakoglobin and beta-catenin: Protein interactions, regulation and biological roles. J. Cell Sci. 2000, 113, 3127–3139. [Google Scholar] [PubMed]
- Liebig, T.; Erasmus, J.; Kalaji, R.; Davies, D.; Loirand, G.; Ridley, A.; Braga, V.M.M. RhoE Is required for keratinocyte differentiation and stratification. Mol. Biol. Cell 2009, 20, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Bektic, J.; Pfeil, K.; Berger, A.P.; Ramoner, R.; Pelzer, A.; Schafer, G.; Kofler, K.; Bartsch, G.; Klocker, H. Small G-protein RhoE is underexpressed in prostate cancer and induces cell cycle arrest and apoptosis. Prostate 2005, 64, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Wajapeyee, N.; Serra, R.W.; Zhu, X.; Mahalingam, M.; Green, M.R. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell 2008, 132, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Yoshie, M.; Hashimoto, K.; Tachikawa, E. Inhibitory effect of insulin-like growth factor-binding protein-7 (IGFBP7) on in vitro angiogenesis of vascular endothelial cells in the rat corpus luteum. J. Reprod. Dev. 2014, 60, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Vishnyakova, T.G.; Kurlander, R.; Bocharov, A.V.; Baranova, I.N.; Chen, Z.; Abu-Asab, M.S.; Tsokos, M.; Malide, D.; Basso, F.; Remaley, A.; et al. CLA-1 and its splicing variant CLA-2 mediate bacterial adhesion and cytosolic bacterial invasion in mammalian cells. Proc. Natl. Acad. Sci. USA 2006, 103, 16888–16893. [Google Scholar] [CrossRef] [PubMed]
- Fritz, G.; Brachetti, C.; Bahlmann, F.; Schmidt, M.; Kaina, B. Rho GTPases in human breast tumours: Expression and mutation analyses and correlation with clinical parameters. Br. J. Cancer 2002, 87, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Adnane, J.; Muro-Cacho, C.; Mathews, L.; Sebti, S.M.; Munoz-Antonia, T. Suppression of rho B expression in invasive carcinoma from head and neck cancer patients. Clin. Cancer Res. 2002, 8, 2225–2232. [Google Scholar] [PubMed]
- Mazieres, J.; Antonia, T.; Daste, G.; Muro-Cacho, C.; Berchery, D.; Tillement, V.; Pradines, A.; Sebti, S.; Favre, G. Loss of RhoB expression in human lung cancer progression. Clin. Cancer Res. 2004, 10, 2742–2750. [Google Scholar] [CrossRef] [PubMed]
- Gambaro, K.; Quinn, M.C.J.; Caceres-Gorriti, K.Y.; Shapiro, R.S.; Provencher, D.; Rahimi, K.; Mes-Masson, A.-M.; Tonin, P.N. Low levels of IGFBP7 expression in high-grade serous ovarian carcinoma is associated with patient outcome. BMC Cancer 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Christenson, L.K.; Gunewardena, S.; Hong, X.; Spitschak, M.; Baufeld, A.; Vanselow, J. Research resource: Preovulatory LH surge effects on follicular theca and granulosa transcriptomes. Mol. Endocrinol. 2013, 27, 1153–1171. [Google Scholar] [CrossRef] [PubMed]
- Burghardt, R.C.; Johnson, G.A.; Jaeger, L.A.; Ka, H.; Garlow, J.E.; Spencer, T.E.; Bazer, F.W. Integrins and extracellular matrix proteins at the maternal-fetal interface in domestic animals. Cells Tissues Organs 2002, 172, 202–217. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.; van Soom, A.; van Zeveren, A.; Favoreel, H.; Peelman, L.J. Quantification of fibronectin 1 (FN1) splice variants, including two novel ones, and analysis of integrins as candidate FN1 receptors in bovine preimplantation embryos. BMC Dev. Biol. 2009, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.; Li, B.; Wang, F.; Tian, C.; Rong, W.; Liu, Y. Identification of differentially expressed genes in Mongolian sheep ovaries by suppression subtractive hybridization. Anim. Reprod. Sci. 2012, 133, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.R.; Fulton, N.; Collins, C.S.; Welsh, M.; Bayne, R.A.L.; Coutts, S.M.; Childs, A.J.; Anderson, R.A. N- and E-cadherin expression in human ovarian and urogenital duct development. Fertil. Steril. 2010, 93, 2348–2353. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Roy, S.K. Expression of E-cadherin and N-cadherin in perinatal hamster ovary: Possible involvement in primordial follicle formation and regulation by follicle-stimulating hormone. Endocrinology 2010, 151, 2319–2330. [Google Scholar] [CrossRef] [PubMed]
- Kodama, T.; Matsukado, Y.; Suzuki, T.; Tanaka, S.; Shimizu, H. Stimulated formation of adenosine 3′,5′-monophosphate by desipramine in brain slices. Biochim. Biophys. Acta 1971, 252, 165–170. [Google Scholar] [CrossRef]
- Mora, J.M.; Fenwick, M.A.; Castle, L.; Baithun, M.; Ryder, T.A.; Mobberley, M.; Carzaniga, R.; Franks, S.; Hardy, K. Characterization and significance of adhesion and junction-related proteins in mouse ovarian follicles. Biol. Reprod. 2012, 86, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Maurya, V.K.; Joshi, A.; Meeran, S.M.; Jha, R.K. Integrin beta 8 (ITGB8) regulates embryo implantation potentially via controlling the activity of TGF-B1 in mice. Biol. Reprod. 2015, 92. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.-L.; Park, A.Y.-J.; Alcaraz, A.; Peng, X.; Jang, I.; Koni, P.; Flavell, R.A.; Gu, H.; Guan, J.-L. Conditional knockout of focal adhesion kinase in endothelial cells reveals its role in angiogenesis and vascular development in late embryogenesis. J. Cell Biol. 2005, 169, 941–952. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, A.; Sato, Y.; Imabayashi, T.; Uemura, T.; Takagi, J.; Sekiguchi, K. Molecular Basis of the Ligand Binding Specificity of alphavbeta8 Integrin. J. Biol. Chem. 2016, 291, 11551–11565. [Google Scholar] [CrossRef] [PubMed]
- Thys, M.; Nauwynck, H.; Maes, D.; Hoogewijs, M.; Vercauteren, D.; Rijsselaere, T.; Favoreel, H.; van Soom, A. Expression and putative function of fibronectin and its receptor (integrin alpha(5)beta(1)) in male and female gametes during bovine fertilization in vitro. Reproduction 2009, 138, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Matsushita, M.; Endo, A.; Kutsukake, M.; Kogo, H. Effect of insulin-like growth factor-binding protein 7 on steroidogenesis in granulosa cells derived from equine chorionic gonadotropin-primed immature rat ovaries. Biol. Reprod. 2007, 77, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Wandji, S.A.; Gadsby, J.E.; Barber, J.A.; Hammond, J.M. Messenger ribonucleic acids for MAC25 and connective tissue growth factor (CTGF) are inversely regulated during folliculogenesis and early luteogenesis. Endocrinology 2000, 141, 2648–2657. [Google Scholar] [CrossRef] [PubMed]
- Casey, O.M.; Fitzpatrick, R.; McInerney, J.O.; Morris, D.G.; Powell, R.; Sreenan, J.M. Analysis of gene expression in the bovine corpus luteum through generation and characterisation of 960 ESTs. Biochim. Biophys. Acta 2004, 1679, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Tsang, P.C.W.; Pate, J.L.; Moses, M.A. A role for cysteine-rich 61 in the angiogenic switch during the estrous cycle in cows: Regulation by prostaglandin F2alpha. Biol. Reprod. 2011, 85, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Sampath, D.; Winneker, R.C.; Zhang, Z. Cyr61, a member of the CCN family, is required for MCF-7 cell proliferation: Regulation by 17beta-estradiol and overexpression in human breast cancer. Endocrinology 2001, 142, 2540–2548. [Google Scholar] [CrossRef] [PubMed]
- MacLaughlan, S.D.; Palomino, W.A.; Mo, B.; Lewis, T.D.; Lininger, R.A.; Lessey, B.A. Endometrial expression of Cyr61: A marker of estrogenic activity in normal and abnormal endometrium. Obstet. Gynecol. 2007, 110, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Borel, F.; Marzocca, F.; Delcros, J.-G.; Rama, N.; Mehlen, P.; Ferrer, J.-L. Molecular characterization of Netrin-1 and APP receptor binding: New leads to block the progression of senile plaques in Alzheimer’s disease. Biochem. Biophys. Res. Commun. 2017, 488, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.W.; Hoshi, T.; Wu, Y.; Sun, X.; Enjyoji, K.; Cszimadia, E.; Sundberg, C.; Robson, S.C. Disordered purinergic signaling inhibits pathological angiogenesis in cd39/Entpd1-null mice. Am. J. Pathol. 2007, 171, 1395–1404. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Huang, W.; Ying, G.; Wu, Q. Sequence analysis and expression mapping of the rat clustered protocadherin gene repertoires. Neuroscience 2007, 144, 579–603. [Google Scholar] [CrossRef] [PubMed]
- Purohit, A.; Sadanandam, A.; Myneni, P.; Singh, R.K. Semaphorin 5A mediated cellular navigation: Connecting nervous system and cancer. Biochim. Biophys. Acta 2014, 1846, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, Y.; Watanabe, K. Contactins: Emerging key roles in the development and function of the nervous system. Cell Adhes. Migr. 2009, 3, 64–70. [Google Scholar] [CrossRef]
- Kidd, T.; Brose, K.; Mitchell, K.J.; Fetter, R.D.; Tessier-Lavigne, M.; Goodman, C.S.; Tear, G. Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell 1998, 92, 205–215. [Google Scholar] [CrossRef]
- Vite, A.; Li, J.; Radice, G.L. New functions for alpha-catenins in health and disease: From cancer to heart regeneration. Cell Tissue Res. 2015, 360, 773–783. [Google Scholar] [CrossRef] [PubMed]
- Sagane, K.; Ohya, Y.; Hasegawa, Y.; Tanaka, I. Metalloproteinase-like, disintegrin-like, cysteine-rich proteins MDC2 and MDC3: Novel human cellular disintegrins highly expressed in the brain. Biochem. J. 1998, 334, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, A.P.; Gossage, S.J.; ffrench-Constant, C. ADAM23 is a cell-surface glycoprotein expressed by central nervous system neurons. J. Neurosci. Res. 2004, 78, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Khan, D.R.; Landry, D.A.; Fournier, E.; Vigneault, C.; Blondin, P.; Sirard, M.-A. Transcriptome meta-analysis of three follicular compartments and its correlation with ovarian follicle maturity and oocyte developmental competence in cows. Physiol. Genom. 2016, 48, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.J.; Ye, Y.; Carroll, A.; Yang, W.; Lee, H.W. Structural biology of the cell adhesion protein CD2: Alternatively folded states and structure-function relation. Curr. Protein Pept. Sci. 2001, 2, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Hattori, N.; Ueda, M.; Fujiwara, H.; Fukuoka, M.; Maeda, M.; Mori, T. Human luteal cells express leukocyte functional antigen (LFA)-3. J. Clin. Endocrinol. Metab. 1995, 80, 78–84. [Google Scholar] [PubMed]
- Soto-Pantoja, D.R.; Kaur, S.; Roberts, D.D. CD47 signaling pathways controlling cellular differentiation and responses to stress. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 212–230. [Google Scholar] [CrossRef] [PubMed]
- Gao, A.G.; Lindberg, F.P.; Finn, M.B.; Blystone, S.D.; Brown, E.J.; Frazier, W.A. Integrin-associated protein is a receptor for the C-terminal domain of thrombospondin. J. Biol. Chem. 1996, 271, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Bornstein, P. Thrombospondins function as regulators of angiogenesis. J. Cell Commun. Signal. 2009, 3, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Berisha, B.; Schams, D.; Rodler, D.; Sinowatz, F.; Pfaffl, M.W. Expression and localization of members of the thrombospondin family during final follicle maturation and corpus luteum formation and function in the bovine ovary. J. Reprod. Dev. 2016, 62, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Roca, J.; Martinez, E.; Vazquez, J.M.; Lucas, X. Selection of immature pig oocytes for homologous in vitro penetration assays with the brilliant cresyl blue test. Reprod. Fertil. Dev. 1998, 10, 479–485. [Google Scholar] [CrossRef] [PubMed]



| Name | Fold | adj.p.Value | ENTREZ Gene ID |
|---|---|---|---|
| ITGB8 | 3.240231 | 1.198584 × 10−2 | 3696 |
| CNTN3 | 3.369908 | 3.018953 × 10−3 | 5067 |
| ITGB1 | 2.730502 | 3.705215 × 10−3 | 3688 |
| ADAM23 | 3.519362 | 7.243130 × 10−3 | 8745 |
| CD9 | 3.036900 | 6.332387 × 10−3 | 928 |
| CD58 | 3.625447 | 4.641977 × 10−3 | 965 |
| LAMA2 | 4.552369 | 7.946265 × 10−4 | 3908 |
| CD47 | 2.054594 | 9.288999 × 10−3 | 961 |
| BMP1 | 3.586853 | 5.391380 × 10−4 | 649 |
| CTNNA2 | 4.349240 | 5.121808 × 10−4 | 1496 |
| RND3 | 4.413617 | 6.618426 × 10−4 | 390 |
| ROBO2 | 3.524243 | 1.183495 × 10−3 | 6092 |
| ENTPD1 | 4.796997 | 3.276430 × 10−4 | 953 |
| PCDH7 | 2.382089 | 6.329369 × 10−4 | 5099 |
| CYR61 | 12.398174 | 7.535476 × 10−5 | 3491 |
| SEMA5A | 2.829721 | 1.092396 × 10−3 | 9037 |
| JUP | 2.264561 | 3.461543 × 10−2 | 3728 |
| RHOB | 2.959342 | 2.898885 × 10−2 | 388 |
| APP | 3.085100 | 5.602323 × 10−3 | 351 |
| LAMB2 | 3.586055 | 1.879115 × 10−4 | 3913 |
| SCARB2 | 2.477578 | 1.415004 × 10−3 | 950 |
| IGFBP7 | 2.476722 | 2.496043 × 10−3 | 3490 |
| TGFBI | 2.282538 | 1.912689 × 10−3 | 7045 |
| Name | Gene Accession Numer | Primer Sequence (5′-3′) | Product Size (bp) |
|---|---|---|---|
| ITGB8 | NM_002214.2 | AAGGGCCAAGTGTGTAGTGG TCTGACATTTGGTCCGCATA | 233 |
| CNTN3 | XM_021069229 | GAATGTTTTGCCCTTGGAAA GCAGCCCATCACTTCTTCTC | 61 |
| ITGB1 | NM_214015.2 | ACCATGCCAATTTCTGCCTGG AACGCACGATCATGTTGGA | 208 |
| ADAM23 | XM_021076147 | GAATCACAGCATGGAAAGCAGT GCATGAGAAGAGCGACAC | 179 |
| CD9 | NM_214006.1 | CAAAGGGACGTACTCTCAAGC GACCCCGAGAAGATGACCAA | 249 |
| CD58 | NM_213795 | CAGTACTGCCAGCGGTGATAT GGAGGCATCGGTAATAAGG | 202 |
| LAMA2 | XM_013992573.1 | GATACAAATGACCCCGTGTTAA TCGAATACAACCTCGGAA | 95 |
| CD47 | NM_213982.1 | TGGAGCCATTCTTTTCATCC AATCAGAAGAGGGCCATGC | 241 |
| BMP1 | XM_021072336.1 | AGCTCTTCGACGGTTACGACA ACAGAATCTCCCGCCGAGT | 93 |
| CTNNA2 | XM_013995995.1 | CCTACCTTCAACGGATTGCCCTT CTGATACTTTGTTGAGGC | 204 |
| RND3 | NM_214296.1 | CCCAACACCAAAATGCTCTTAA GTGGCTGCTCCAATCTGT | 145 |
| ROBO2 | XM_013982523.1 | GGAACAGCTTCTTCTAAGGGAA TAAAGAAATTGTTCATTGCACT | 238 |
| ENTPD1 | NM_214153.1 | GCTATGGGAAGGATCAAGCAGT GCAGGGAGCCTCATAAAG | 139 |
| PCDH7 | NM_001244484.1 | TTCCACTCCCAGAGGACAACG GTCAGGGCTACATCTGGAA | 83 |
| CYR61 | XM_001927740.4 | CCAATGACAACCCCGACTGCCC GGTACTTCTTCACGCTGG | 176 |
| SEMA5A | XM_013984924.1 | AACACCAGCATAACCAACCACA ACTGGGGAATTACAAGAAGC | 221 |
| JUP | NM_214323.1 | ATCCCATGGACACCTACAGCGG CTCAGGCACTTTGCTATC | 148 |
| RHOB | NM_001123189.1 | TATGTGCTTCTCGGTGGACACG AGGTAGTCGTAGGCTTGG | 230 |
| APP | NM_214372 | TGGGGAAAGACACAAACCCTT CATGCACTAGTTTGATACAGCTT | 206 |
| LAMB2 | XM_013981664.1 | GCTGCCCAAGGATGACCACAT CCTCCTGTTCGCACTAGCTT | 130 |
| SCARB2 | NM_001244155.1 | AGTCGCCTGAAGTCTGTGGT AGTTGCCCCATGTCGTAGTC | 236 |
| IGFBP7 | NM_001163801.1 | ATAACCTGGCCATTCAGACG ACAGCTCAGCACCTTCACCT | 207 |
| TGFBI | NM_214015.2 | ACCATGCCAATTTCTGCCTGGA ACGCACGATCATGTTGGA | 208 |
| PBGD | NM_001097412.1 | GAGAGTGCCCCTATGATGCTAT GATGGCACTGAACTCCT | 214 |
| ACTB | XM_003124280.3 | CCCTTGCCGCTCCGCCTTCGCA GCAATATCGGTCATCCAT | 69 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Budna, J.; Celichowski, P.; Bryja, A.; Dyszkiewicz-Konwińska, M.; Jeseta, M.; Bukowska, D.; Antosik, P.; Brüssow, K.P.; Bruska, M.; Nowicki, M.; et al. Significant Down-Regulation of “Biological Adhesion” Genes in Porcine Oocytes after IVM. Int. J. Mol. Sci. 2017, 18, 2685. https://doi.org/10.3390/ijms18122685
Budna J, Celichowski P, Bryja A, Dyszkiewicz-Konwińska M, Jeseta M, Bukowska D, Antosik P, Brüssow KP, Bruska M, Nowicki M, et al. Significant Down-Regulation of “Biological Adhesion” Genes in Porcine Oocytes after IVM. International Journal of Molecular Sciences. 2017; 18(12):2685. https://doi.org/10.3390/ijms18122685
Chicago/Turabian StyleBudna, Joanna, Piotr Celichowski, Artur Bryja, Marta Dyszkiewicz-Konwińska, Michal Jeseta, Dorota Bukowska, Paweł Antosik, Klaus Peter Brüssow, Małgorzata Bruska, Michał Nowicki, and et al. 2017. "Significant Down-Regulation of “Biological Adhesion” Genes in Porcine Oocytes after IVM" International Journal of Molecular Sciences 18, no. 12: 2685. https://doi.org/10.3390/ijms18122685







