Applications of Alternative Nucleases in the Age of CRISPR/Cas9
Abstract
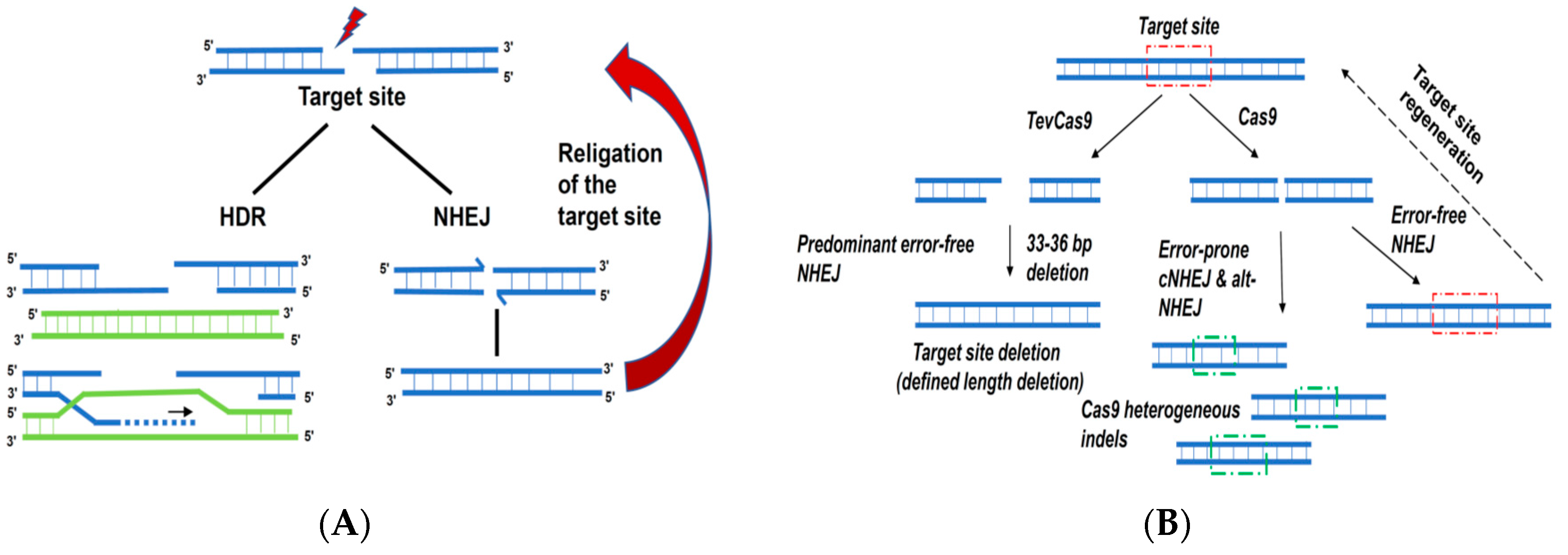
:1. Introduction
2. Genome-Editing Reagents Based on Nuclease Domains That Dimerize
2.1. ZFNs: Zinc-Finger Nucleases
2.2. TALENs: Transcription Activator-Like Effector Nucleases
2.3. Other Dimeric Nucleases
3. Monomeric Nucleases
3.1. Monomeric Nucleases with Unrelated Binding and Cleavage Domains
3.1.1. Single-Chain Variants of ZFNs and TALENs
3.1.2. GIY-YIG-Derived Nucleases
3.2. Single-Chain Monomeric Endonucleases
3.2.1. Meganucleases and MegaTALs
3.2.2. CRISPR/Cas9
4. Regulating Nuclease Activity
5. Biasing Genome-Editing Events
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jiang, F.; Doudna, J.A. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Burstein, D.; Harrington, L.; Strutt, S.C.; Probst, A.J.; Anantharaman, K.; Thomas, B.C.; Doudna, J.A.; Banfield, J.F. New CRISPR-Cas systems from uncultivated microbes. Nature 2017, 542, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfora, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Chandrasegaran, S.; Carroll, D. Origins of programmable nucleases for genome engineering. J. Mol. Biol. 2016, 428, 963–989. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D. Genome engineering with targetable nucleases. Annu. Rev. Biochem. 2014, 83, 409–439. [Google Scholar] [CrossRef] [PubMed]
- Mak, A.N.; Bradley, P.; Bogdanove, A.J.; Stoddard, B.L. TAL effectors: Function, structure, engineering and applications. Curr. Opin. Struct. Biol. 2013, 23, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, B.L. Homing endonucleases: From microbial genetic invaders to reagents for targeted DNA modification. Structure 2011, 19, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.; Poirot, L.; Galetto, R.; Smith, J.; Montoya, G.; Duchateau, P.; Pâques, F. Meganucleases and other tools for targeted genome engineering: Perspectives and challenges for gene therapy. Curr. Gene Ther. 2011, 11, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.J.; Diez-Villasenor, C.; Garcia-Martinez, J.; Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009, 155, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Staals, R.H.; Zhu, Y.; Taylor, D.W.; Kornfeld, J.E.; Sharma, K.; Barendregt, A.; Koehorst, J.J.; Vlot, M.; Neupane, N.; Varossieau, K.; et al. RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol. Cell 2014, 56, 518–530. [Google Scholar] [CrossRef] [PubMed]
- Tamulaitis, G.; Kazlauskiene, M.; Manakova, E.; Venclovas, Č.; Nwokeoji, A.O.; Dickman, M.J.; Horvath, P.; Siksnys, V. Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol. Cell 2014, 56, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Normile, D. China sprints ahead in CRISPR therapy race. Science 2017, 358, 20–21. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wu, L.P.; Chandrasegaran, S. Functional domains in Fok I restriction endonuclease. Proc. Natl. Acad. Sci. USA 1992, 89, 4275–4279. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Bibikova, M.; Whitby, F.G.; Reddy, A.R.; Chandrasegaran, S.; Carroll, D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000, 28, 3361–3369. [Google Scholar] [CrossRef] [PubMed]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Porteus, M.H.; Carroll, D. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 2005, 23, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Moscou, M.J.; Bogdanove, A.J. A simple cipher governs DNA recognition by TAL effectors. Science 2009, 326, 1501. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Mak, A.N.; Bradley, P.; Cernadas, R.A.; Bogdanove, A.J.; Stoddard, B.L. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 2012, 335, 716–719. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Yan, C.; Pan, X.; Mahfouz, M.; Wang, J.; Zhu, J.-K.; Shi, Y.; Yan, N. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science 2012, 335, 720–723. [Google Scholar] [CrossRef] [PubMed]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.C.; Tan, S.; Qiao, G.; Barlow, K.A.; Wang, J.; Xia, D.F.; Meng, X.; Paschon, D.E.; Leung, E.; Hinkley, S.J.; et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011, 29, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Huang, S.; Jiang, W.Z.; Wright, D.; Spalding, M.H.; Weeks, D.P.; Yang, B. TAL nucleases (TALNs): Hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011, 39, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Lamb, B.M.; Mercer, A.C.; Barbas, C.F., III. Directed evolution of the TALE N-terminal domain for recognition of all 5′ bases. Nucleic Acids Res. 2013, 41, 9779–9785. [Google Scholar] [CrossRef] [PubMed]
- Doyle, E.L.; Hummel, A.W.; Demorest, Z.L.; Starker, C.G.; Voytas, D.F.; Bradley, P.; Bogdanove, A.J. TAL effector specificity for base 0 of the DNA target is altered in a complex, effector- and assay-dependent manner by substitutions for the tryptophan in cryptic repeat-1. PLoS ONE 2013, 8, e82120. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Y.; Wu, X.; Wang, J.; Wang, Y.; Qiu, Z.; Chang, T.; Huang, H.; Lin, R.J.; Yee, J.K. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat. Biotechnol. 2015, 33, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Holkers, M.; Maggio, I.; Liu, J.; Janssen, J.M.; Miselli, F.; Mussolino, C.; Recchia, A.; Cathomen, T.; Gonçalves, M.A. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013, 41, e63. [Google Scholar] [CrossRef] [PubMed]
- Stella, S.; Molina, R.; López-Méndez, B.; Juillerat, A.; Bertonati, C.; Daboussi, F.; Campos-Olivas, R.; Duchateau, P.; Montoya, G. BuD, a helix-loop-helix DNA-binding domain for genome modification. Acta Crystallogr. D 2014, 70, 2042–2052. [Google Scholar] [CrossRef] [PubMed]
- Schierling, B.; Dannemann, N.; Gabsalilow, L.; Wende, W.; Cathomen, T.; Pingoud, A. A novel zinc-finger nuclease platform with a sequence-specific cleavage module. Nucleic Acids Res. 2012, 40, 2623–2638. [Google Scholar] [CrossRef] [PubMed]
- Schierling, B.; Wende, W.; Pingoud, A. Redesigning the single-chain variant of the restriction endonuclease PvuII by circular permutation. FEBS Lett. 2012, 586, 1736–1741. [Google Scholar] [CrossRef] [PubMed]
- Fonfara, I.; Curth, U.; Pingoud, A.; Wende, W. Creating highly specific nucleases by fusion of active restriction endonucleases and catalytically inactive homing endonucleases. Nucleic Acids Res. 2012, 40, 847–860. [Google Scholar] [CrossRef] [PubMed]
- Guilinger, J.P.; Thompson, D.B.; Liu, D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014, 32, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Aouida, M.; Eid, A.; Ali, Z.; Cradick, T.; Lee, C.; Deshmukh, H.; Atef, A.; AbuSamra, D.; Gadhoum, S.Z.; Merzaban, J.; et al. Efficient fdCas9 synthetic endonuclease with improved specificity for precise genome engineering. PLoS ONE 2015, 10, e0133373. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Q.; Wyvekens, N.; Khayter, C.; Foden, J.A.; Thapar, V.; Reyon, D.; Goodwin, M.J.; Aryee, M.J.; Joung, J.K. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014, 32, 569–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, N.; Zhao, H. A single-chain TALEN architecture for genome engineering. Mol. Biosyst. 2014, 10, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Minczuk, M.; Papworth, M.A.; Miller, J.C.; Murphy, M.P.; Klug, A. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Res. 2008, 36, 3926–3938. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Wolfs, J.M.; Kolaczyk, T.; Roberts, A.K.; Hu, S.X.; Edgell, D.R. Monomeric site-specific nucleases for genome editing. Proc. Natl. Acad. Sci. USA 2012, 109, 8061–8066. [Google Scholar] [CrossRef] [PubMed]
- Wolfs, J.M.; DaSilva, M.; Meister, S.E.; Wang, X.; Schild-Poulter, C.; Edgell, D.R. MegaTevs: Single-chain dual nucleases for efficient gene disruption. Nucleic Acids Res. 2014, 42, 8816–8829. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Wang, L.; Wolfs, J.A.; Kolaczyk, T.; McDowell, B.; Wang, X.; Schild-Poulter, C.; Bogdanove, A.J.; Edgell, D.R. The I-TevI Nuclease and Linker Domains Contribute to the Specificity of Monomeric TALENs. G3 Bethesda 2014, 4, 1155–1165. [Google Scholar] [CrossRef] [PubMed]
- Wolfs, J.M.; Hamilton, T.A.; Lant, J.T.; Laforet, M.; Zhang, J.; Salemi, L.M.; Gloor, G.B.; Schild-Poulter, C.; Edgell, D.R. Biasing genome-editing events toward precise length deletions with an RNA-guided TevCas9 dual nuclease. Proc. Natl. Acad. Sci. USA 2016, 113, 14988–14993. [Google Scholar] [CrossRef] [PubMed]
- Beurdeley, M.; Bietz, F.; Li, J.; Thomas, S.; Stoddard, T.; Juillerat, A.; Zhang, F.; Voytas, D.F.; Duchateau, P.; Silva, G.H. Compact designer TALENs for efficient genome engineering. Nat. Commun. 2013, 4, 1762–1768. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Wolfs, J.M.; Edgell, D.R. The monomeric GIY-YIG homing endonuclease I-BmoI uses a molecular anchor and a flexible tether to sequentially nick DNA. Nucleic Acids Res. 2013, 41, 5413–5427. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.R.; Hallinan, J.P.; Shen, B.W.; Chik, J.K.; Bolduc, J.M.; Kulshina, N.; Robins, L.I.; Kaiser, B.K.; Jarjour, J.; Havens, K.; et al. Indirect DNA sequence recognition and its impact on nuclease cleavage activity. Structure 2016, 24, 862–873. [Google Scholar] [CrossRef] [PubMed]
- Molina, R.; Redondo, P.; Stella, S.; Marenchino, M.; D’Abramo, M.; Gervasio, F.L.; Charles Epinat, J.; Valton, J.; Grizot, S.; Duchateau, P.; et al. Non-specific protein-DNA interactions control I-CreI target binding and cleavage. Nucleic Acids Res. 2012, 40, 6936–6945. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, R.; Choi, M.; Stoddard, B.L. Redesign of extensive protein-DNA interfaces of meganucleases using iterative cycles of in vitro compartmentalization. Proc. Natl. Acad. Sci. USA 2014, 111, 4061–4066. [Google Scholar] [CrossRef] [PubMed]
- Thyme, S.B.; Boissel, S.J.; Arshiya, S.Q.; Nolan, T.; Baker, D.A.; Park, R.U.; Kusak, L.; Ashworth, J.; Baker, D. Reprogramming homing endonuclease specificity through computational design and directed evolution. Nucleic Acids Res. 2014, 42, 2564–2576. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Grizot, S.; Arnould, S.; Duclert, A.; Epinat, J.C.; Chames, P.; Prieto, J.; Redondo, P.; Blanco, F.J.; Bravo, J.; et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006, 34, e149. [Google Scholar] [CrossRef] [PubMed]
- Sather, B.D.; Romano-Ibarra, G.S.; Sommer, K.; Curinga, G.; Hale, M.; Khan, I.F.; Singh, S.; Song, Y.; Gwiazda, K.; Sahni, J.; et al. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci. Transl. Med. 2015, 7, 307ra156. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Khan, I.F.; Boissel, S.; Jarjour, J.; Pangallo, J.; Thyme, S.; Baker, D.; Scharenberg, A.M.; Rawlings, D.J. Progressive engineering of a homing endonuclease genome editing reagent for the murine X-linked immunodeficiency locus. Nucleic Acids Res. 2014, 42, 6463–6475. [Google Scholar] [CrossRef] [PubMed]
- Aubert, M.; Boyle, N.M.; Stone, D.; Stensland, L.; Huang, M.L.; Magaret, A.S.; Galetto, R.; Rawlings, D.J.; Scharenberg, A.M.; Jerome, K.R. In vitro inactivation of latent HSV by targeted mutagenesis using an HSV-specific homing endonuclease. Mol. Ther. Nucleic Acids 2014, 3, e146. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, I.G.; Prieto, J.; Subramanian, S.; Coloma, J.; Redondo, P.; Villate, M.; Merino, N.; Marenchino, M.; D’abramo, M.; Gervasio, F.L.; et al. Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus. Nucleic Acids Res. 2011, 39, 729–743. [Google Scholar] [CrossRef] [PubMed]
- Redondo, P.; Prieto, J.; Muñoz, I.G.; Alibés, A.; Stricher, F.; Serrano, L.; Cabaniols, J.P.; Daboussi, F.; Arnould, S.; Perez, C.; et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature 2008, 456, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Boissel, S.; Jarjour, J.; Astrakhan, A.; Adey, A.; Gouble, A.; Duchateau, P.; Shendure, J.; Stoddard, B.L.; Certo, M.T.; Baker, D.; et al. megaTALs: A rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 2014, 42, 2591–2601. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Zhang, L.; Zhou, X.; Chen, X.; Huang, G.; Li, F.; Wang, R.; Wu, N.; Yan, Y.; Tong, C.; et al. Induction of site-specific chromosomal translocations in embryonic stem cells by CRISPR/Cas9. Sci. Rep. 2016, 6, 21918. [Google Scholar] [CrossRef] [PubMed]
- Blasco, R.B.; Karaca, E.; Ambrogio, C.; Cheong, T.C.; Karayol, E.; Minero, V.G.; Voena, C.; Chiarle, R. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep. 2014, 9, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Koo, T.; Lee, J.; Kim, J.-S. Measuring and Reducing Off-Target Activities of Programmable Nucleases Including CRISPR-Cas9. Mol. Cells 2015, 38, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Ramirez, C.L.; Linder, S.J.; Pattanayak, V.; Shoresh, N.; Ku, M.; Foden, J.A.; Reyon, D.; Bernstein, B.E.; Liu, D.R.; et al. In silico abstraction of zinc finger nuclease cleavage profiles reveals an expanded landscape of off-target sites. Nucleic Acids Res. 2013, 41, e181. [Google Scholar] [CrossRef] [PubMed]
- Pattanayak, V.; Ramirez, C.L.; Joung, J.K.; Liu, D.R. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods 2011, 8, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Meng, X.; Zhu, L.J.; Lawson, N.D.; Wolfe, S.A. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011, 39, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, R.; Lombardo, A.; Arens, A.; Miller, J.C.; Genovese, P.; Kaeppel, C.; Nowrouzi, A.; Bartholomae, C.C.; Wang, J.; Friedman, G.; et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat. Biotechnol. 2011, 29, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Szczepek, M.; Brondani, V.; Büchel, J.; Serrano, L.; Segal, D.J.; Cathomen, T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 2007, 25, 786–793. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Dagdas, Y.S.; Kleinstiver, B.P.; Welch, M.M.; Sousa, A.A.; Harrington, L.B.; Sternberg, S.H.; Joung, J.K.; Yildiz, A.; Doudna, J.A. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature 2017, 550, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Lin, C.-Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Molina, R.; Marcaida, M.J.; Redondo, P.; Marenchino, M.; Duchateau, P.; D’Abramo, M.; Montoya, G.; Prieto, J. Engineering a nickase on the homing endonuclease I-DmoI scaffold. J. Biol. Chem. 2015, 290, 18534–18544. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, C.L.; Certo, M.T.; Mussolino, C.; Goodwin, M.J.; Cradick, T.J.; McCaffrey, A.P.; Cathomen, T.; Scharenberg, A.M.; Joung, J.K. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res. 2012, 40, 5560–5568. [Google Scholar] [CrossRef] [PubMed]
- Ousterout, D.G.; Gersbach, C.A. The Development of TALE Nucleases for Biotechnology. Methods Mol. Biol. 2016, 1338, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Zetsche, B.; Volz, S.E.; Zhang, F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015, 33, 139–142. [Google Scholar] [CrossRef] [PubMed]
- Nihongaki, Y.; Kawano, F.; Nakajima, T.; Sato, M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 2015, 33, 755–760. [Google Scholar] [CrossRef] [PubMed]
- Mandegar, M.A.; Huebsch, N.; Frolov, E.B.; Shin, E.; Truong, A.; Olvera, M.P.; Chan, A.H.; Miyaoka, Y.; Holmes, K.; Spencer, C.I.; et al. CRISPR interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell Stem Cell 2016, 18, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Guha, T.K.; Wai, A.; Hausner, G. Programmable genome editing tools and their regulation for efficient genome engineering. Comput. Struct. Biotechnol. J. 2017, 15, 146–160. [Google Scholar] [CrossRef] [PubMed]
- Pawluk, A.; Davidson, A.R.; Maxwell, K.L. Anti-CRISPR: Discovery, mechanism and function. Nat. Rev. Microbiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Harrington, L.B.; Doxzen, K.W.; Ma, E.; Liu, J.J.; Knott, G.J.; Edraki, A.; Garcia, B.; Amrani, N.; Chen, J.S.; Cofsky, J.C.; et al. A broad-spectrum inhibitor of CRISPR-Cas9. Cell 2017, 170, 1224–1233. [Google Scholar] [CrossRef] [PubMed]
- Pawluk, A.; Amrani, N.; Zhang, Y.; Garcia, B.; Hidalgo-Reyes, Y.; Lee, J.; Edraki, A.; Shah, M.; Sontheimer, E.J.; Maxwell, K.L.; et al. Naturally occurring off-Switches for CRISPR-Cas9. Cell 2016, 167, 1829–1838. [Google Scholar] [CrossRef] [PubMed]
- Rauch, B.J.; Silvis, M.R.; Hultquist, J.F.; Waters, C.S.; McGregor, M.J.; Krogan, N.J.; Bondy-Denomy, J. Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell 2017, 168, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Pawluk, A.; Staals, R.H.; Taylor, C.; Watson, B.N.; Saha, S.; Fineran, P.C.; Maxwell, K.L.; Davidson, A.R. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat. Microbiol. 2016, 1, 16085. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, T.; Nakade, S.; Sakane, Y.; Suzuki, K.T.; Yamamoto, T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat. Protoc. 2016, 11, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Daer, R.M.; Cutts, J.P.; Brafman, D.A.; Haynes, K.A. The impact of chromatin dynamics on Cas9-mediated genome editing in human cells. ACS Synth. Biol. 2017, 6, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Rinsma, M.; Janssen, J.M.; Liu, J.; Maggio, I.; Gonçalves, M.A. Probing the impact of chromatin conformation on genome editing tools. Nucleic Acids Res. 2016, 44, 6482–6492. [Google Scholar] [CrossRef] [PubMed]
- Horlbeck, M.A.; Witkowsky, L.B.; Guglielmi, B.; Replogle, J.M.; Gilbert, L.A.; Villalta, J.E.; Torigoe, S.E.; Tjian, R.; Weissman, J.S. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. eLife 2016, 5, e12677. [Google Scholar] [CrossRef] [PubMed]
- Hinz, J.M.; Laughery, M.F.; Wyrick, J.J. Nucleosomes inhibit Cas9 endonuclease activity in vitro. Biochemistry 2015, 54, 7063–7066. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Haemmerle, M.; Genovese, G.; Draetta, G.F.; Chin, L. Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Rep. 2016, 14, 1555–1566. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Staahl, B.T.; Alla, R.K.; Doudna, J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 2014, 3, e04766. [Google Scholar] [CrossRef] [PubMed]
- Certo, M.T.; Gwiazda, K.S.; Kuhar, R.; Sather, B.; Curinga, G.; Mandt, T.; Brault, M.; Lambert, A.R.; Baxter, S.K.; Jacoby, K.; et al. Coupling endonucleases with DNA end-processing enzymes to drive gene disruption. Nat. Methods 2012, 9, 973–975. [Google Scholar] [CrossRef] [PubMed]
- Delacôte, F.; Perez, C.; Guyot, V.; Duhamel, M.; Rochon, C.; Ollivier, N.; Macmaster, R.; Silva, G.H.; Pâques, F.; Daboussi, F.; et al. High frequency targeted mutagenesis using engineered endonucleases and DNA-end processing enzymes. PLoS ONE 2013, 8, e53217. [Google Scholar] [CrossRef] [PubMed]
- Richardson, C.D.; Ray, G.J.; DeWitt, M.A.; Curie, G.L.; Corn, J.E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016, 34, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Till, B.J.; Burtner, C.; Comai, L.; Henikoff, S. Mismatch cleavage by single-strand specific nucleases. Nucleic Acids Res. 2004, 32, 2632–2641. [Google Scholar] [CrossRef] [PubMed]
- Vouillot, L.; Thélie, A.; Pollet, N. Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. Genes Genomes Genet. 2015, 5, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Qiu, P.; Shandilya, H.; D’Alessio, J.M.; O’Connor, K.; Durocher, J.; Gerard, G.F. Mutation detection using Surveyor nuclease. Biotechniques 2004, 36, 702–707. [Google Scholar] [PubMed]
- Maruyama, T.; Dougan, S.K.; Truttmann, M.C.; Bilate, A.M.; Ingram, J.R.; Ploegh, H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015, 33, 420–538. [Google Scholar] [CrossRef] [PubMed]
- Miyaoka, Y.; Berman, J.R.; Cooper, S.B.; Mayerl, S.J.; Chan, A.J.; Zhang, B.; Karlin-Neumann, G.A.; Conklin, B.R. Systematic quantification of HDR and NHEJ reveals effects of locus, nuclease, and cell type on genome-editing. Sci. Rep. 2016, 6, 23549. [Google Scholar] [CrossRef] [PubMed]
- Certo, M.T.; Ryu, B.Y.; Annis, J.E.; Garibov, M.; Jarjour, J.; Rawlings, D.J.; Scharenberg, A.M. Tracking genome engineering outcome at individual DNA breakpoints. Nat. Methods 2011, 8, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Zaboikin, M.; Zaboikina, T.; Freter, C.E.; Srinivasakumar, N. Differentiation of Genome edited human iPSCs into endothelial progenitor cells for hemophilia A therapy. Blood 2016, 128, 1474. [Google Scholar]
- Urnov, F.D.; Miller, J.C.; Lee, Y.L.; Beausejour, C.M.; Rock, J.M.; Augustus, S.; Jamieson, A.C.; Porteus, M.H.; Gregory, P.D.; Holmes, M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 2005, 435, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Geisinger, J.M.; Turan, S.; Hernandez, S.; Spector, L.P.; Calos, M.P. In vivo blunt-end cloning through CRISPR/Cas9-facilitated non-homologous end-joining. Nucleic Acids Res. 2016, 44, e76. [Google Scholar] [CrossRef] [PubMed]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a Class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Petersen, T.S.; Jensen, K.T.; Bolund, L.; Kühn, R.; Luo, Y. Fusion of SpCas9 to E. coli Rec A protein enhances CRISPR-Cas9 mediated gene knockout in mammalian cells. J. Biotechnol. 2017, 247, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Dolgin, E. CRISPR hacks enable pinpoint repairs to genome. Nature 2017, 550, 439–440. [Google Scholar] [CrossRef] [PubMed]
- Hess, G.T.; Tycko, J.; Yao, D.; Bassik, M.C. Methods and applications of CRISPR-mediated base editing in eukaryotic genomes. Mol. Cell 2017, 68, 26–43. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Nishida, K.; Arazoe, T.; Yachie, N.; Banno, S.; Kakimoto, M.; Tabata, M.; Mochizuki, M.; Miyabe, A.; Araki, M.; Hara, K.Y.; et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 2016, 353. [Google Scholar] [CrossRef] [PubMed]
- Vojta, A.; Dobrinić, P.; Tadić, V.; Bočkor, L.; Korać, P.; Julg, B.; Klasić, M.; Zoldoš, V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016, 44, 5615–5628. [Google Scholar] [CrossRef] [PubMed]
- Hilton, I.B.; D’Ippolito, A.M.; Vockley, C.M.; Thakore, P.; Crawford, G.E.; Reddy, T.E.; Gersbach, C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015, 33, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.S.; Wu, H.; Ji, X.; Seltzer, Y.; Wu, X.; Czauderna, S.; Shu, J.; Dadon, D.; Young, R.A.; Jaenisch, R. Editing DNA methylation in the mammalian genome. Cell 2016, 167, 233–247. [Google Scholar] [CrossRef] [PubMed]
| Nuclease Domain | Property | Type of DSB | Associated Reagents |
|---|---|---|---|
| FokI | Type IIS, dimeric, non-specific nuclease | 3-nt 5′ overhang | ZFNs, TALENs, Cas9-FokI |
| I-TevI | GIY-YIG, monomeric, site-specific | 2-nt 3′ overhang | ZFEs, TALENs, MegaTev, TevCas9 |
| PvuII | Type IIP, homodimeric, site-specific | Blunt end | I-SceI-PvuII, ZF-PvuII, PvuII-LHE |
| Recombinase | Serine recombinase (Sin recombinase); invertase Gin | Not applicable | ZF-recombinase, TALE-recombinase |
| Cas9 | Type II CRISPR/Cas family; RuvC/HNH nuclease domains; monomeric; requires PAM sequence; moderate specificity | Blunt end | CRISPR/Cas9, CRISPRi, Cas9-FokI |
| Cpf1 | Monomeric; non-specific; recognizes T-rich PAM sequence at the 5′ of the guide RNA | 5-nt 5′ overhang | CRISPR/Cpf1 |
| Meganuclease | LAGLIDADG family, monomeric or dimeric; very specific | 4-nt 3′ overhang | MegaTAL, TALE-I-SceI, MegaTev |
| Property | Cas9 | ZFN | TALEN | Meganuclease |
|---|---|---|---|---|
| Specificity (off-target) | Relatively non-specific | Relatively non-specific | Specific | Very specific |
| Biasing events (repair) | NHEJ | NHEJ | HDR | HDR |
| Design & targeting constraints | PAM requirement (NGG for SpCas9) | Context-dependent assembly of ZFs; GC rich targets preferred | Assembly of TALE repeats; 5′ targeted base is T | Re-design of protein-DNA interface; central 4 bases intolerant to change |
| Dimerization required | No | Yes | Yes | No |
| Coding sequence | Long | Short | Long and repetitive | Short |
| Therapeutic delivery | Easy | Moderate | Moderate | Easy |
| Vector packaging | Moderate | Difficult | Difficult | Easy |
| Multiplex potential | High | Low | Low | High |
| Cost-effective | Yes | No | Moderate | No |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guha, T.K.; Edgell, D.R. Applications of Alternative Nucleases in the Age of CRISPR/Cas9. Int. J. Mol. Sci. 2017, 18, 2565. https://doi.org/10.3390/ijms18122565
Guha TK, Edgell DR. Applications of Alternative Nucleases in the Age of CRISPR/Cas9. International Journal of Molecular Sciences. 2017; 18(12):2565. https://doi.org/10.3390/ijms18122565
Chicago/Turabian StyleGuha, Tuhin K., and David R. Edgell. 2017. "Applications of Alternative Nucleases in the Age of CRISPR/Cas9" International Journal of Molecular Sciences 18, no. 12: 2565. https://doi.org/10.3390/ijms18122565





