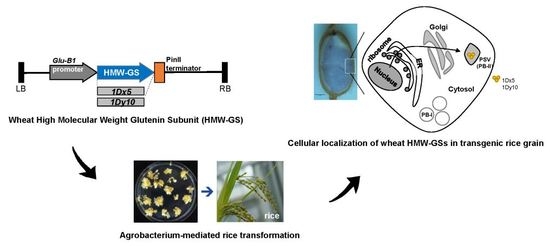
Cellular Localization of Wheat High Molecular Weight Glutenin Subunits in Transgenic Rice Grain
Abstract
:1. Introduction
2. Results and Discussion
2.1. High-Molecular-Weight Glutenin Subunits (HMW-GSs) Amino Acid Sequence Analysis
2.2. Selection of Transgenic Rice Plants Expressing Wheat HMW-GSs
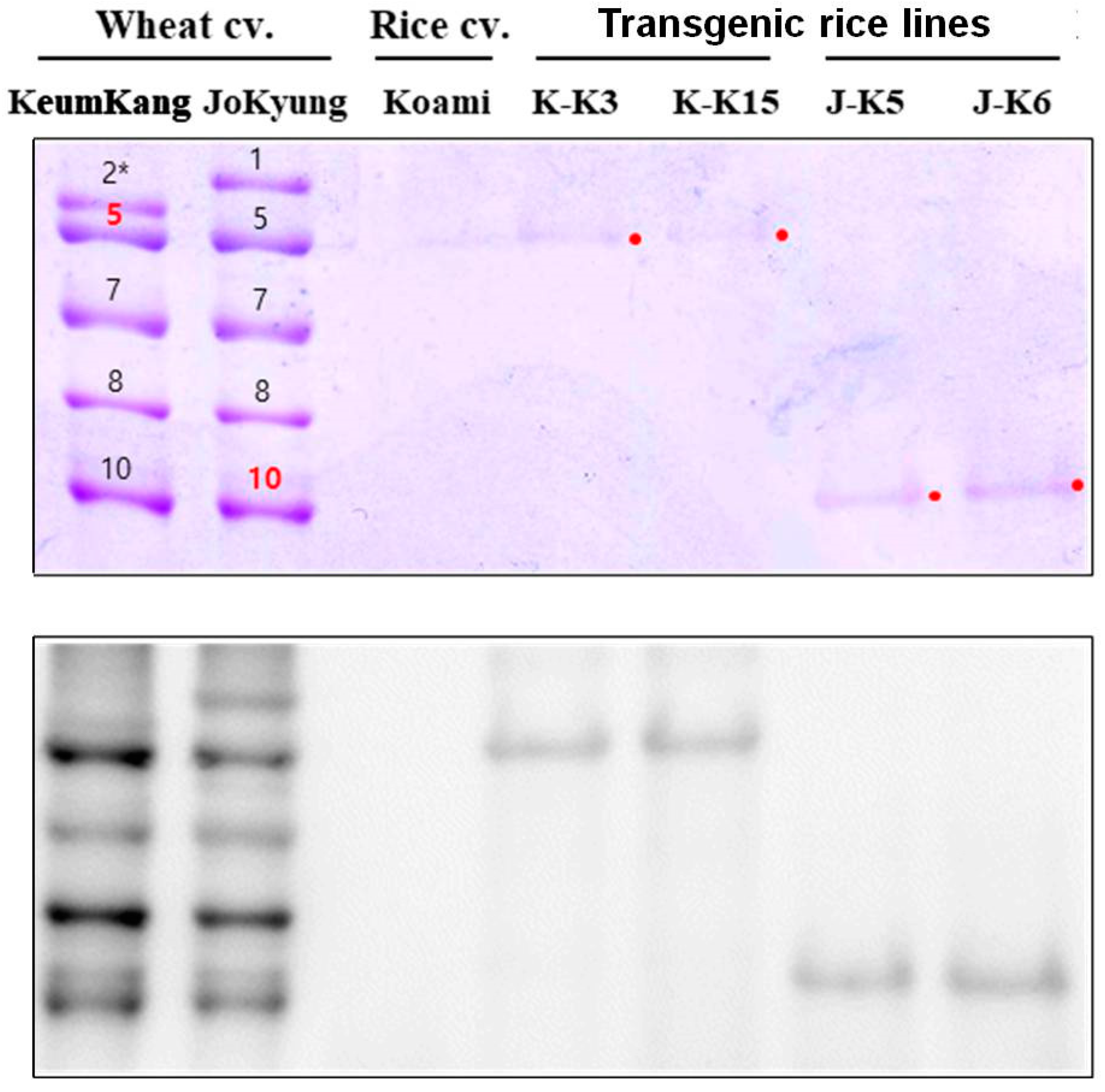
2.3. Accumulation of Wheat HMW-GSs in Transgenic Rice Endosperms
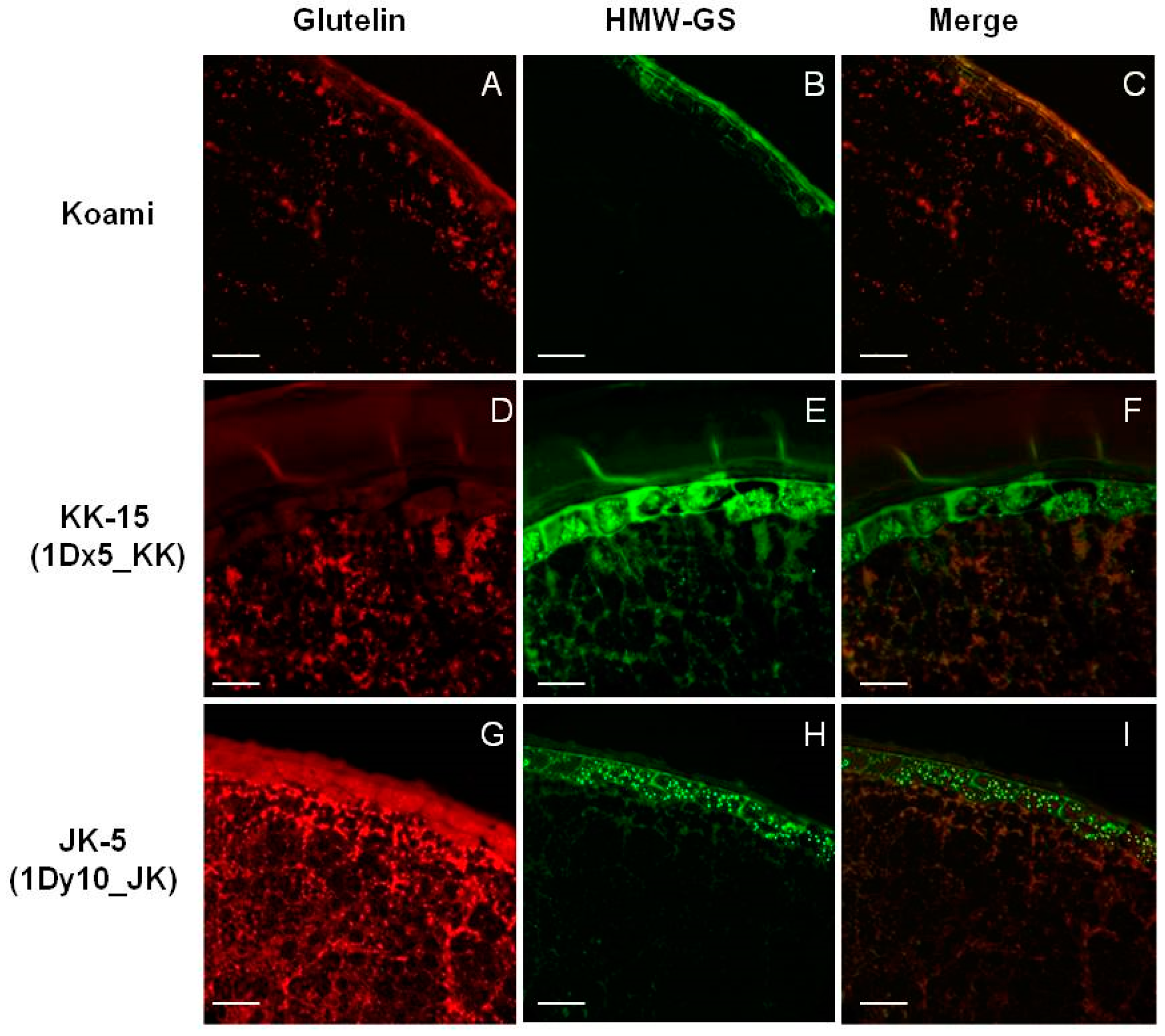
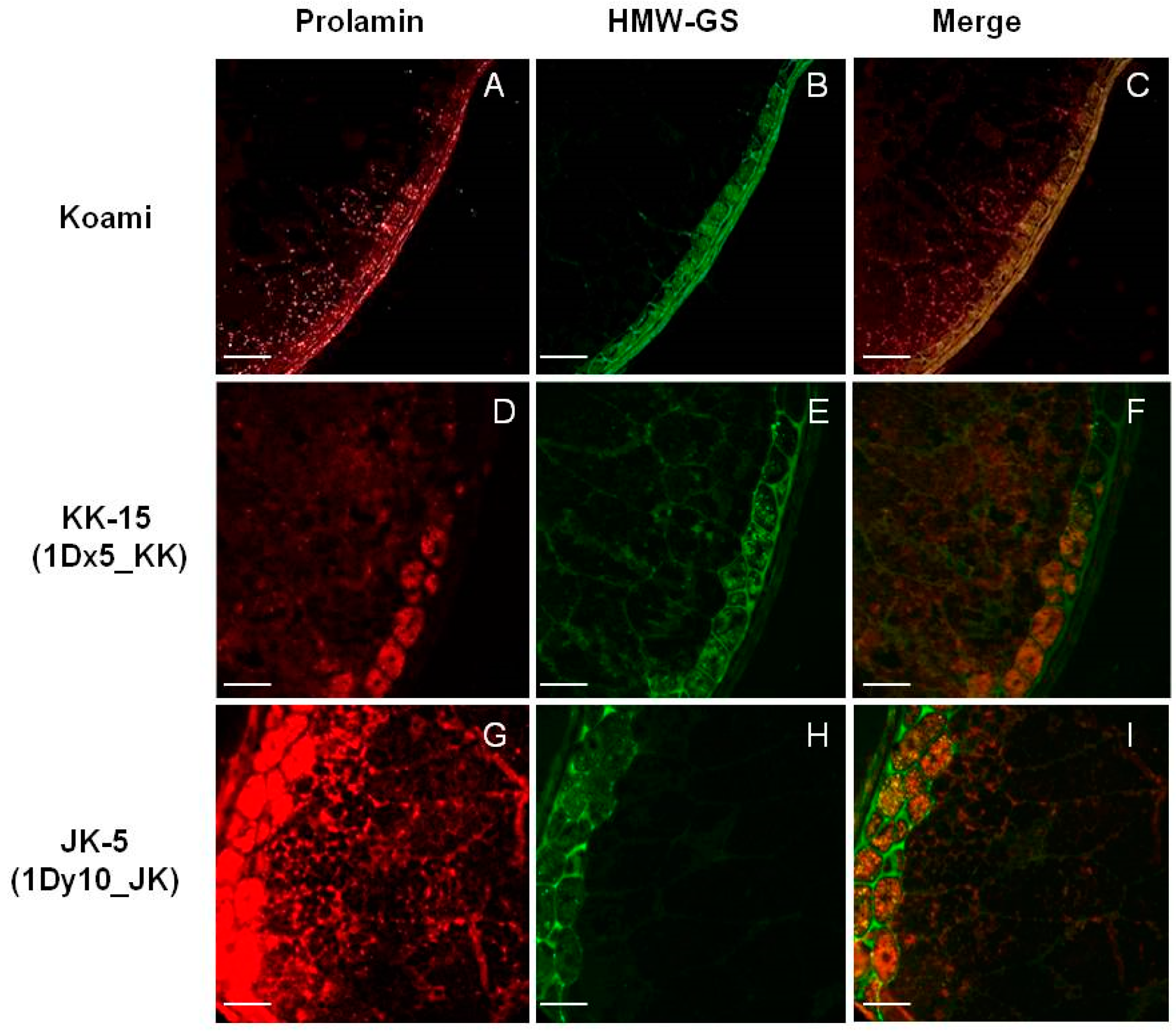
2.4. Cellular Localization of Wheat HMW-GSs in Transgenic Rice Seed Endosperm
3. Materials and Methods
3.1. Vector Construction and Rice Transformation
3.2. Antibodies
3.3. Quantitative Real Time-Polymerase Chain Reaction (qRT-PCR)
3.4. SDS-PAGE and Immunoblotting
3.5. In Situ Immunohybridization
3.6. Immunofluorescence
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kawakatsu, T.; Hirose, S.; Yasuda, H.; Takaiwa, F. Reducing rice seed storage protein accumulation leads to changes in nutrient quality and storage organelle formation. Plant Physiol. 2010, 154, 1842–1854. [Google Scholar] [CrossRef] [PubMed]
- Shewry, P.R.; Halford, N.G. Cereal seed storage proteins: Structures, properties and role in grain utilization. J. Exp. Bot. 2002, 53, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Payne, P.I. Genetics of wheat storage proteins and the effect of allelic variation on bread-making quality. Annu. Rev. Plant Physiol. 1987, 38, 141–153. [Google Scholar]
- Halford, N.G.; Forde, J.; Anderson, O.D.; Greene, F.C.; Shewry, P.R. The nucleotide and deduced amino acid sequences of an HMW glutenin subunit gene from chromosome 1B of bread wheat (Triticum aestivum L.) and comparison with those of genes from chromosomes 1A and 1D. Theor. Appl. Genet. 1987, 75, 117–126. [Google Scholar] [CrossRef]
- Anjum, F.M.; Khan, M.R.; Din, A.; Saeed, M.; Pasha, I.; Arshad, M.U. Wheat gluten: High molecular weight glutenin subunits-structure, genetics, and relation to dough elasticity. J. Food Sci. 2007, 72, R56–R63. [Google Scholar] [CrossRef] [PubMed]
- Galili, G.; Sengupta-Gopalan, C.; Ceriotti, A. The endoplasmic reticulum of plant cells and its role in protein maturation and biogenesis of oil bodies. Plant Mol. Biol. 1998, 38, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Loussert, C.; Popineau, Y.; Mangavel, C. Protein bodies ontogeny and localization of prolamin components in the developing endosperm of wheat caryopses. J. Cereal Sci. 2008, 47, 445–456. [Google Scholar]
- Tosi, P.; Parker, M.; Gritsch, C.S.; Carzaniga, R.; Martin, B.; Shewry, P.R. Trafficking of storage proteins in developing grain of wheat. J. Exp. Bot. 2009, 60, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, H.; Tanaka, K. The site of synthesis and accumulation of rice storage proteins. Plant Cell Physiol. 1986, 27, 135–145. [Google Scholar]
- Kawakatsu, T.; Yamamoto, M.P.; Hirose, S.; Yano, M.; Takaiwa, F. Characterization of a new rice glutelin gene GluD-1 expressed in the starchy endosperm. J. Exp. Bot. 2008, 59, 4233–4245. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.H.; Messing, J. Amplification of prolamin storage protein genes in different subfamilies of the Poaceae. Theor. Appl. Genet. 2009, 119, 1397–1412. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, Y.; Coughlan, S.J.; Okita, T.W.; Satoh, H.; Ogawa, M.; Kumamaru, T. The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol. 2002, 128, 1212–1222. [Google Scholar] [CrossRef] [PubMed]
- Van der Borght, A.; Vandeputte, G.E.; Derycke, V.; Brijs, K.; Daenen, G.; Delcour, J.A. Extractability and chromatographic separation of rice endosperm proteins. J. Cereal Sci. 2006, 44, 68–74. [Google Scholar] [CrossRef]
- Saito, Y.; Shigemitsu, T.; Yamasaki, R.; Sasou, A.; Goto, F.; Kishida, K.; Kuroda, M.; Tanaka, K.; Morita, S.; Satoh, S.; et al. Formation mechanism of the internal structure of type I protein bodies in rice endosperm: Relationship between the localization of prolamin species and the expression of individual genes. Plant 2012, 70, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Kreis, M.B.; Forde, G.; Rahman, S.; Miflin, B.J.; Shewry, P.R. Molecular evolution of the seed storage proteins of barley, rye and wheat. J. Mol. Biol. 1985, 183, 499–502. [Google Scholar] [CrossRef]
- Okita, T.W.; Cheesbrough, V.; Reeves, C.D. Evolution and heterogeneity of the α-/β-type and γ-type gliadin DNA sequences. J. Biol. Chem. 1985, 260, 8203–8213. [Google Scholar] [PubMed]
- Cameron-Mills, V.; Brandt, A. A γ-hordein gene. Plant Mol. Biol. 1988, 11, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Shyur, L.F.; Wen, T.N.; Chen, C.S. Purification and characterization of rice prolamins. Bot. Bull. Acad. Sin. 1994, 35, 65–71. [Google Scholar]
- Koehler, P.; Wieser, H. Chemistry of cereal grains. In Handbook on Sourdough Biotechnology; Springer: New York, NY, USA, 2013; pp. 11–45. [Google Scholar]
- Herman, E.M.; Larkins, B.A. Protein storage bodies and vacuoles. Plant Cell 1999, 11, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Shigemitsu, T.; Tanaka, K.; Morita, S.; Satoh, S.; Masumura, T. Ultrastructure of mature protein body in the starchy endosperm of dry cereal grain. Biosci. Biotechnol. Biochem. 2010, 74, 1485–1487. [Google Scholar] [CrossRef] [PubMed]
- Motoyama, T.; Maruyama, N.; Amari, Y.; Kobayashi, K.; Washida, H.; Higasa, T.; Takaiwa, F.; Utsumi, S. α’s Subunit of soybean β-conglycinin forms complex with rice glutelin via a disulphide bond in transgenic rice seeds. J. Exp. Bot. 2009, 60, 4015–4027. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Hiroi, T.; Hirose, S.; Yang, L.; Takaiwa, F. Rice seed ER-derived protein body as an efficient delivery vehicle for oral tolerogenic peptides. Peptides 2010, 31, 1421–1425. [Google Scholar] [CrossRef] [PubMed]
- Sivaramakrishnan, H.P.; Senge, B.; Chattopadhyay, P.K. Rheological properties of rice dough for making rice bread. J. Food Eng. 2004, 62, 37–45. [Google Scholar] [CrossRef]
- Shin, M. Rice-processed food. Food Sci. Ind. 2009, 42, 2–18. [Google Scholar]
- Rooke, L.; Bekes, F.; Fido, R.; Barro, F.; Gras, P.; Tatham, A.S.; Barcelo, P.; Lazzeri, P.; Shewry, P.R. Overexpression of a gluten protein in transgenic wheat results in greatly increased dough strength. J. Cereal Sci. 1999, 30, 115–120. [Google Scholar] [CrossRef]
- Popineau, Y.; Deshayes, G.; Lefebvre, J.; Fido, R.; Tatham, A.S.; Shewry, P.R. Prolamin aggregation, gluten viscoelasticity, and mixing properties of transgenic wheat lines expressing 1Ax and 1Dx high molecular weight glutenin subunit transgenes. J. Agric. Food Chem. 2001, 49, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Gadaleta, A.; Blechl, A.E.; Nguyen, S.; Cardone, M.F.; Ventura, M.; Quick, J.S.; Blanco, A. Stably expressed d-genome-derived HMW glutenin subunit genes transformed into different durum wheat genotypes change dough mixing properties. Mol. Breed. 2008, 22, 267–279. [Google Scholar] [CrossRef]
- Altpeter, F.; Juan, C.P.; Wieser, H. Stable expression of 1Dx5 and 1Dy10 high-molecular-weight glutenin subunit genes in transgenic rye drastically increases the polymeric glutelin fraction in rye flour. Plant Mol. Biol. 2004, 54, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Jeong, N.; Jeon, S.H.; Kim, D.Y.; Lee, C.; Ok, H.C.; Park, K.D.; Hong, H.C.; Lee, S.S.; Moon, J.K.; Park, S.K. Development of marker-free TaGlu-Ax1 transgenic rice harboring a wheat high-molecular-weight glutenin Subunit (HMW-GS) protein. J. Life Sci. 2016, 26, 1121–1129. [Google Scholar] [CrossRef]
- Park, S.K.; Shin, D.; Hwang, W.H.; Oh, S.Y.; Cho, J.H.; Han, S.I.; Nam, M.H.; Park, D.S. Development of marker-free transgenic rice for increasing bread-making quality using wheat high molecular weight glutenin subunits (HMW-GS) gene. J. Life Sci. 2013, 23, 1317–1324. [Google Scholar] [CrossRef]
- Oszvald, M.; Jenes, B.; Tomoskozi, S.; Bekes, F.; Tamas, L. Expression of the 1Dx5 high molecular weight glutenin subunit protein in transgenic rice. Cereal Res. Commun. 2007, 35, 1543–1549. [Google Scholar]
- Oszvald, M.; Balazs, G.; Polya, S.; Tomoskozi, S.; Appels, R.; Bekes, F.; Tamas, L. Wheat storage proteins in transgenic rice endosperm. J. Agric. Food Chem. 2013, 61, 7606–7614. [Google Scholar] [CrossRef] [PubMed]
- Park, S.K.; Shin, D.; Hwang, W.H.; Hur, Y.J.; Kim, T.H.; Oh, S.Y.; Cho, J.H.; Han, S.I.; Lee, S.S.; Nam, M.H. Development of marker-free transgenic rice expressing the wheat storage protein, Glu-1Dy10, for increasing quality processing of bread and noodles. J. Life Sci. 2014, 24, 618–625. [Google Scholar] [CrossRef]
- Qu, L.Q.; Xing, Y.P.; Liu, W.X.; Xu, X.P.; Song, Y.R. Expression pattern and activity of six glutelin gene promoters in transgenic rice. J. Exp. Bot. 2008, 59, 2417–2424. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.R.; Beom, H.R.; Altenbach, S.B.; Lee, M.K.; Lim, S.H.; Lee, J.Y. Improved method for reliable HMW-GS identification by RP-HPLC and SDS-PAGE in common wheat cultivars. Molecules 2017, 22, 1055. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, A.; Xia, X.C.; Yan, Y.M.; Appels, R.; Mahmood, T.; He, Z.H. Wheat seed storage proteins: Advances in molecular genetics, diversity and breeding applications. J. Cereal Sci. 2014, 60, 11–24. [Google Scholar] [CrossRef]
- He, Z.H.; Liu, L.; Xia, X.C.; Liu, J.J.; Pena, R.J. Composition of HMW and LMW glutenin subunits and their effects on dough properties, pan bread, and noodle quality of Chinese bread wheats. Cereal Chem. 2005, 82, 345–350. [Google Scholar] [CrossRef]
- Choi, S.B.; Wang, C.; Muench, D.G.; Ozawa, K.; Franceschi, V.R.; Wu, Y.J.; Okita, T.W. Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 2000, 407, 765–767. [Google Scholar] [PubMed]
- Yamagata, H.; Sugimoto, T.; Tanaka, K.; Kasai, Z. Biosynthesis of storage proteins in developing rice seeds. Plant Physiol. 1982, 70, 1094–1100. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Jo, Y.M.; Lee, J.Y.; Lim, S.H.; Kim, Y.M. Lack of globulin synthesis during seed development alters accumulation of seed storage proteins in rice. Int. J. Mol. Sci. 2015, 16, 14717–14736. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Guo, F.; Liu, B.; Huang, N.; Watkins, S.C. Expression and localization of human lysozyme in the endosperm of transgenic rice. Planta 2003, 216, 597–603. [Google Scholar] [PubMed]
- Yang, L.; Suzuki, K.; Hirose, S.; Wakasa, T.; Takaiwa, F. Development of transgenic rice seed accumulating a major Japanese cedar pollen allergen (Cry j 1) structurally disrupted for oral immunotherapy. Plant Biotechnol. J. 2007, 5, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Kishida, K.; Takata, K.; Takahashi, H.; Shimada, T.; Tanaka, K.; Morita, S.; Satoh, S.; Masumura, T. A green fluorescent protein fused to rice prolamin forms protein body-like structures in transgenic rice. J. Exp. Bot. 2009, 60, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Shigemitsu, T.; Masumura, T.; Morita, S.; Satoh, S. Accumulation of rice prolamin-GFP fusion proteins induces ER-derived protein bodies in transgenic rice calli. Plant Cell Rep. 2013, 32, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Inze, D.; Depicker, A. GATEWAY((TM)) vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Kim, Y.M.; Lee, J.Y.; Yoon, Y.H.; Choi, S.B.; Ha, S.H.; Lim, S.H. New design of rice seed storage proteins. J. Plant Biotechnol. 2011, 38, 263–271. [Google Scholar] [CrossRef]
- Cho, K.; Shibato, J.; Kubo, A.; Kohno, Y.; Satoh, K.; Kikuchi, S.; Agrawal, G.K.; Sarkar, A.; Rakwal, R. Genome-wide mapping of the ozone-responsive transcriptomes in rice panicle and seed tissues reveals novel insight into their regulatory events. Biotechnol. Lett. 2013, 35, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Shepherd, K.; Cornish, G. A simplified SDS-PAGE procedure for separating LMW subunits of glutenin. J. Cereal Sci. 1991, 14, 203–208. [Google Scholar] [CrossRef]



© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jo, Y.-M.; Cho, K.; Lee, H.-J.; Lim, S.-H.; Kim, J.S.; Kim, Y.-M.; Lee, J.-Y. Cellular Localization of Wheat High Molecular Weight Glutenin Subunits in Transgenic Rice Grain. Int. J. Mol. Sci. 2017, 18, 2458. https://doi.org/10.3390/ijms18112458
Jo Y-M, Cho K, Lee H-J, Lim S-H, Kim JS, Kim Y-M, Lee J-Y. Cellular Localization of Wheat High Molecular Weight Glutenin Subunits in Transgenic Rice Grain. International Journal of Molecular Sciences. 2017; 18(11):2458. https://doi.org/10.3390/ijms18112458
Chicago/Turabian StyleJo, Yeong-Min, Kyoungwon Cho, Hye-Jung Lee, Sun-Hyung Lim, Jin Sun Kim, Young-Mi Kim, and Jong-Yeol Lee. 2017. "Cellular Localization of Wheat High Molecular Weight Glutenin Subunits in Transgenic Rice Grain" International Journal of Molecular Sciences 18, no. 11: 2458. https://doi.org/10.3390/ijms18112458