Current Status of Early Blight Resistance in Tomato: An Update
Abstract
:1. Introduction
2. Early Blight
2.1. Causal Agent
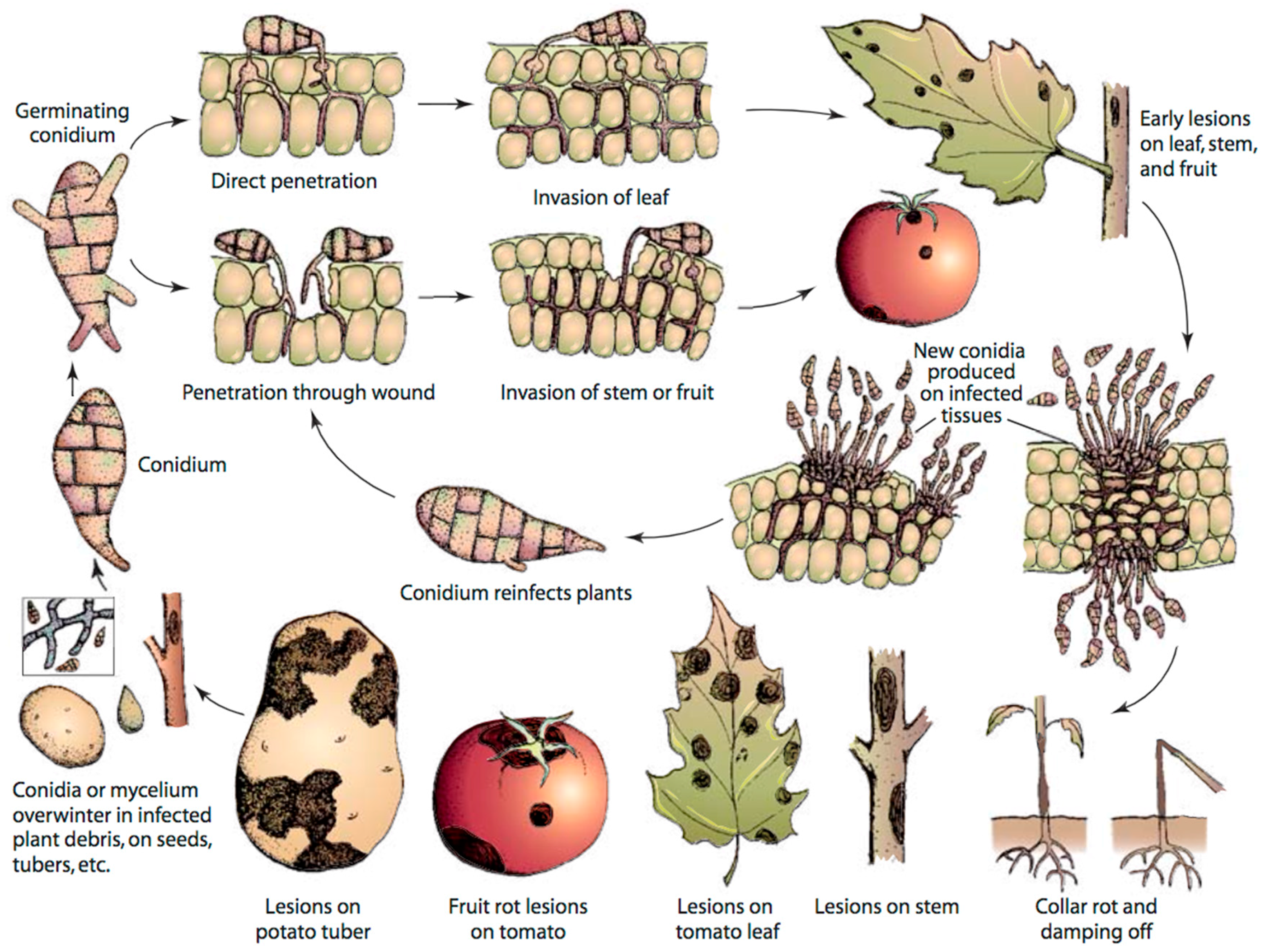
2.2. Disease Cycle
2.3. Variability among Isolates
2.4. Molecular Mechanism of Infection
2.5. Disease Symptoms
2.6. Control Measures
3. Screening Methods
3.1. Inoculum Preparation
3.2. Field Screening
3.3. Greenhouse Screening
3.4. Laboratory Assays (Detached Leaflet Assay)
4. Genetic Source of Resistance
Characterization of EB Resistance
5. Breeding for EB Resistance
6. Conclusions and Future Prospects
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Foolad, M. Genome mapping and molecular breeding of tomato. Int. J. Plant Genom. 2007. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT Database. Available online: http://faostat.fao.org/site/339/default.aspx (accessed on 19 September 2017).
- Bohm, F.; Edge, R.; Truscott, G. Interactions of dietary carotenoids with activated (singlet) oxygen and free radicals: Potential effects for human health. Mol. Nutr. Food Res. 2012, 56, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Van Breemen, R.B.; Pajkovic, N. Multitargeted therapy of cancer by lycopene. Cancer Lett. 2008, 269, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.P.; Khan, A.; Nino-Liu, D.; Foolad, M.R. A molecular linkage map of tomato displaying chromosomal locations of resistance gene analogs based on a Lycopersicon esculentum × Lycopersicon hirsutum cross. Genome 2002, 45, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Rotem, J. The Genus Alternaria: Biology, Epidemiology, and Pathogenicity; The American Phtyopathological Society: St. Paul, MN, USA, 1994; Volume 326, p. 48. [Google Scholar]
- Simmons, E.G. Alternaria themes and variations (244–286) species on Solanaceae. Mycotaxon 2000, 75, 1–115. [Google Scholar]
- Woudenberg, J.H.C.; Truter, M.; Groenewald, J.Z.; Crous, P.W. Large-spored Alternaria pathogens in section Porri disentangled. Stud. Mycol. 2014, 79, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Bessadat, N.; Berruyer, R.; Hamon, B.; Bataille-Simoneau, N.; Benichou, S.; Kihal, M.; Henni, D.E.; Simoneau, P. Alternaria species associated with early blight epidemics on tomato and other Solanaceae crops in northwestern Algeria. Eur. J. Plant Pathol. 2017, 148, 181–197. [Google Scholar] [CrossRef] [Green Version]
- Gannibal, P.B.; Orina, A.S.; Mironenko, N.V.; Levitin, M.M. Differentiation of the closely related species, Alternaria solani and A. tomatophila, by molecular and morphological features and aggressiveness. Eur. J. Plant Pathol. 2014, 139, 609–623. [Google Scholar] [CrossRef]
- Pavon, M.A.; Luna, A.; de la Cruz, S.; Gonzalez, I.; Martin, R.; Garcia, T. PCR-based assay for the detection of Alternaria species and correlation with HPLC determination of altenuene, alternariol and alternariol monomethyl ether production in tomato products. Food Control 2012, 25, 45–52. [Google Scholar] [CrossRef]
- Edin, E. Species specific primers for identification of Alternaria solani, in combination with analysis of the F129L substitution associates with loss of sensitivity toward strobilurins. Crop Prot. 2012, 38, 72–73. [Google Scholar] [CrossRef]
- Kumar, S.; Singh, R.; Kashyap, P.L.; Srivastava, A.K. Rapid detection and quantification of Alternaria solani in tomato. Sci. Hortic. 2013, 151, 184–189. [Google Scholar] [CrossRef]
- Perez-Martinez, S.; Snowdon, R.; Pons-Kuhnemann, J. Variability of Cuban and international populations of Alternaria solani from different hosts and localities: AFLP genetic analysis. Eur. J. Plant Pathol. 2004, 110, 399–409. [Google Scholar] [CrossRef]
- Andersen, B.; Dongo, A.; Pryor, B.M. Secondary metabolite profiling of Alternaria dauci, A. porri, A. solani, and A. tomatophila. Mycol. Res. 2008, 112, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Brun, S.; Madrid, H.; Van den Ende, B.G.; Andersen, B.; Marinach-Patrice, C.; Mazier, D.; De Hoog, G.S. Multilocus phylogeny and MALDI-TOF analysis of the plant pathogenic species Alternaria dauci and relatives. Fungal Biol. 2013, 117, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.X.; Pryor, B.; Peever, T.; Lawrence, C.B. The Alternaria genomes database: A comprehensive resource for a fungal genus comprised of saprophytes, plant pathogens, and allergenic species. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Foolad, M.R.; Merk, H.L.; Ashrafi, H. Genetics, genomics and breeding of late blight and early blight resistance in tomato. Crit. Rev. Plant Sci. 2008, 27, 75–107. [Google Scholar] [CrossRef]
- Jones, J.P. Compendium of Tomato Diseases. In Early Blight; Jones, J.B., Jones, J.P., Stall, R.E., Zitter, T.A., Eds.; APS Press: St. Paul, MN, USA, 1991. [Google Scholar]
- Kemmitt, G. Early blight of potato and tomato. In The Plant Health Instructor; The American Phytopathological Society (APS): St. Paul, MN, USA, 2002. [Google Scholar]
- Sherf, A.F.; Macnab, A.A. Vegetable Diseases and Their Control; John Wiley and Sons: New York, NY, USA, 1986; p. 728. [Google Scholar]
- Agrios, G.N. Plant Pathology; Elsevier Academic Press: San Diego, CA, USA, 2005; Volume 5. [Google Scholar]
- Chaerani, R.; Voorrips, R. Tomato early blight (Alternaria solani): The pathogen, genetics, and breeding for resistance. J. Gen. Plant Pathol. 2006, 72, 335–347. [Google Scholar] [CrossRef]
- Pasche, J.S.; Wharam, C.M.; Gudmestad, N.C. Shift in sensitivity of Alternaria solani in response to QoI fungicides. Plant Dis. 2004, 88, 181–187. [Google Scholar] [CrossRef]
- Chaerani, R.; Smulders, M.J.M.; van der Linden, C.G.; Vosman, B.; Stam, P.; Voorrips, R.E. QTL identification for early blight resistance (Alternaria solani) in a Solanum lycopersicum × S. arcanum cross. Theor. Appl. Genet. 2007, 114, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Akagi, Y.; Akamatsu, H.; Otani, H.; Kodama, M. Horizontal Chromosome Transfer, a Mechanism for the Evolution and Differentiation of a Plant-Pathogenic Fungus. Eukaryot. Cell 2009, 8, 1732–1738. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B. Alternaria spp.: From general saprophyte to specific parasite. Mol. Plant Pathol. 2003, 4, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Tsuge, T.; Harimoto, Y.; Akimitsu, K.; Ohtani, K.; Kodama, M.; Akagi, Y.; Egusa, M.; Yamamoto, M.; Otani, H. Host-Selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol. Rev. 2013, 37, 44–66. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, K.; Miyamoto, T.; Fujimoto, T.; Oguri, H.; Tokiwano, T.; Oikawa, H.; Ebizuka, Y.; Fujii, I. Solanapyrone Synthase, a Possible Diels-Alderase and Iterative Type I Polyketide Synthase Encoded in a Biosynthetic Gene Cluster from Alternaria solani. ChemBioChem 2010, 11, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Park, C.M.; Park, J.J.; Akamatsu, H.O.; Peever, T.L.; Xian, M.; Gang, D.R.; Vandemark, G.; Chen, W.D. Functional Analyses of the Diels-Alderase Gene sol5 of Ascochyta rabiei and Alternaria solani Indicate that the Solanapyrone Phytotoxins Are Not Required for Pathogenicity. Mol. Plant Microbe Int. 2015, 28, 482–496. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, M.; Thangavelu, B.; Chun, S.C.; Sathiyabama, M. Proteases from phytopathogenic fungi and their importance in phytopathogenicity. J. Gen. Plant Pathol. 2016, 82, 233–239. [Google Scholar] [CrossRef]
- Chandrasekaran, M.; Chandrasekar, R.; Chun, S.C.; Sathiyabama, M. Isolation, characterization and molecular three-dimensional structural predictions of metalloprotease from a phytopathogenic fungus, Alternaria solani (Ell. and Mart.) Sor. J. Biosci. Bioeng. 2016, 122, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, M.; Chandrasekar, R.; Sa, T.; Sathiyabama, M. Serine protease identification (in vitro) and molecular structure predictions (in silico) from a phytopathogenic fungus, Alternaria solani. J. Basic Microbiol. 2014, 54, S210–S218. [Google Scholar] [CrossRef] [PubMed]
- Collinge, D.B.; Kragh, K.M.; Mikkelsen, J.D.; Nielsen, K.K.; Rasmussen, U.; Vad, K. Plant chitinases. Plant J. 1993, 3, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Nasir, I.A.; Tabassum, B.; Aaliya, K.; Tariq, M.; Rao, A.Q. Expression studies of chitinase gene in transgenic potato against Alternaria solani. Plant Cell Tissue Organ. Cult. 2017, 128, 563–576. [Google Scholar] [CrossRef]
- Upadhyay, P.; Ganie, S.H.; Rai, A.; Singh, M.; Sinha, B. Identification of transcription factors in tomato, potentially related to early blight resistance at invasion in host tissue, using microarray expression profiling. S. Afr. J. Bot. 2016, 106, 165–173. [Google Scholar] [CrossRef]
- Rosenzweig, N.; Atallah, Z.K.; Olaya, G.; Stevenson, W.R. Evaluation of QoI fungicide application strategies for managing fungicide resistance and potato early blight epidemics in Wisconsin. Plant Dis. 2008, 92, 561–568. [Google Scholar] [CrossRef]
- Leiminger, J.H.; Adolf, B.; Hausladen, H. Occurrence of the F129L mutation in Alternaria solani populations in Germany in response to QoI application, and its effect on sensitivity. Plant Pathol. 2014, 63, 640–650. [Google Scholar] [CrossRef]
- Barksdale, T.H.; Stoner, A.K. Study of inheritance of tomato early blight resistance. Plant Dis. Rep. 1977, 61, 63–70. [Google Scholar]
- Ivors, K.L.; Louws, F.J. 2013 North Carolina Agricultural Chemicals Manual. In College of Agriculture and Life Sciences; North Carolina State University: Raleigh, NC, USA, 2013. [Google Scholar]
- Joseph, A.; Igbinosa, O.B.; Alori, E.T.; Ademiluyi, B.O.; Aluko, A.P. Effectiveness of Pseudomonas species in the management of tomato early blight pathogen Alternaria solani. Afr. J. Microbiol. Res. 2017, 11, 972–976. [Google Scholar]
- Jagadeesh, K.S.; Jagadeesh, D.R. Biological Control of Early Blight of Tomato Caused by Alternaria solani as Influenced by Different Delivery Methods of Pseudomonas gladioli B25. In II International Symposium on Tomato Diseases; Saygili, H., Sahin, F., Aysan, Y., Eds.; ISHS: Kusadasi, Turkey, 2009; Volume 808, pp. 327–332. [Google Scholar]
- Yeole, G.J.; Teli, N.P.; Kotkar, H.M.; Mendki, P.S. Cinnamomum zeylanicum extracts and their formulations control early blight of tomato. J. Biopestic. 2014, 7, 110. [Google Scholar]
- Sarkar, S.; Beura, S.K.; Nandi, A.; Das, S.; Dash, S.K.; Senapati, N.; Patnaik, A. Management of Early blight of tomato (Alternaria solani Ellis and Martin) by chemicals and biocontrol agents under field condition. J. Mycopathol. Res. 2016, 54, 81–84. [Google Scholar]
- Chowdappa, P.; Kumar, S.P.M.; Lakshmi, M.J.; Upreti, K.K. Growth stimulation and induction of systemic resistance in tomato against early and late blight by Bacillus subtilis OTPB1 or Trichoderma harzianum OTPB3. Biol. Control 2013, 65, 109–117. [Google Scholar] [CrossRef]
- Pane, C.; Zaccardelli, M. Evaluation of Bacillus strains isolated from solanaceous phylloplane for biocontrol of Alternaria early blight of tomato. Biol. Control 2015, 84, 11–18. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Rybakova, D.; Grube, M.; Koberl, M. The plant microbiome explored: Implications for experimental botany. J. Exp. Bot. 2016, 67, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Bordenstein, S.R.; Theis, K.R. Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes. PLoS Biol. 2015, 13, e1002226. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Jousset, A. Plant Breeding Goes Microbial. Trends Plant Sci. 2017, 22, 555–558. [Google Scholar] [CrossRef] [PubMed]
- Kroll, S.; Agler, M.T.; Kemen, E. Genomic dissection of host-microbe and microbe-microbe interactions for advanced plant breeding. Curr. Opin. Plant Biol. 2017, 36, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Bakker, M.G.; Manter, D.K.; Sheflin, A.M.; Weir, T.L.; Vivanco, J.M. Harnessing the rhizosphere microbiome through plant breeding and agricultural management. Plant Soil 2012, 360, 1–13. [Google Scholar] [CrossRef]
- Locke, S.B. A method for measuring resistance to defoliation disease in tomato and other Lycopersicon species. Phytopathology 1948, 38, 937–942. [Google Scholar]
- Vakalounakis, D.J. Control of early blight of greenhouse tomato, caused by Alternaria solani, by inhibiting sporulation with ultraviolet-absorbing vinyl film. Plant Dis. 1991, 75, 795–797. [Google Scholar] [CrossRef]
- Maiero, M.; Ng, T.J.; Barksdale, T.H. Combining ability estimates for early blight resistance in tomato. J. Am. Soc. Hortic. Sci. 1989, 114, 118–121. [Google Scholar]
- Shahin, E.A.; Shepard, J.F. An efficient technique for inducing profuse sporulation of Alternaria species. Phytopatholy 1979, 69, 618–620. [Google Scholar] [CrossRef]
- Barksdale, T.H. Resistance of tomato seedling to early blight. Phytopathology 1969, 59, 443. [Google Scholar]
- Foolad, M.R.; Ntahimpera, N.; Christ, B.J.; Lin, G.Y. Comparison of field, greenhouse, and detached-leaflet evaluations of tomato germ plasm for early blight resistance. Plant Dis. 2000, 84, 967–972. [Google Scholar] [CrossRef]
- Foolad, M.R.; Subbiah, P.; Ghangas, G.S. Parent-offspring correlation estimate of heritability for early blight resistance in tomato, Lycopersicon esculentum Mill. Euphytica 2002, 126, 291–297. [Google Scholar] [CrossRef]
- Horsfall, J.G.; Barratt, R.W. An improved grading system for measuring plant diseases. Phytopathology 1945, 35, 655. [Google Scholar]
- Nash, A.F.; Gardner, R.G. Heritability of tomato early blight resistance derived from Lycopersicon hirsutum PI 126445. J Am. Soc. Hortic. Sci. 1988, 113, 264–268. [Google Scholar]
- Nash, A.F.; Gardner, R.G. Tomato early blight resistance in a breeding line derived from Lycopersicon hirsutum PI 126445. Plant Dis. 1988, 72, 206–209. [Google Scholar] [CrossRef]
- Gardner, R.G. Use of Lycopersicon-hirsutum PI 126445 in breeding early blight-resistant tomatoes. Hortscience 1984, 19, 208. [Google Scholar]
- Gardner, R.G. Greenhouse disease screen facilitates breeding resistance to tomato early blight. Hortscience 1990, 25, 222–223. [Google Scholar]
- Vloutoglou, I.; Kalogerakis, S.N. Effects of inoculum concentration, wetness duration and plant age on development of early blight (Alternaria solani) and on shedding of leaves in tomato plants. Plant Pathol. 2000, 49, 339–345. [Google Scholar] [CrossRef]
- Gardner, R.G. NC-EBR-1 and NC-EBR-2 Early blight resistant tomato breeding lines. Hortscience 1988, 23, 779–781. [Google Scholar]
- Gardner, R.G. ‘Plum Dandy’, a hybrid tomato, and its parents, NC EBR-5 and NC EBR-6. Hortscience 2000, 35, 962–963. [Google Scholar]
- Gardner, R.G.; Shoemaker, P.B. ‘Mountain supreme’ early blight-resistant hybrid tomato and its parents, NC EBR-3 and NC EBR-4. Hortscience 1999, 34, 745–746. [Google Scholar]
- Panthee, D.R.; Gardner, R.G. ‘Mountain Merit’: A late blight-resistant large-fruited tomato hybrid. HortScience 2010, 45, 1547–1548. [Google Scholar]
- Gardner, R.G.; Panthee, D.R. ‘Mountain Magic’: An Early Blight and Late Blight-Resistant Specialty Type F-1 Hybrid Tomato. Hortscience 2012, 47, 299–300. [Google Scholar]
- Andrus, C.F.; Reynard, G.B.; Jorgensen, H.; Eades, J. Collar rot resistance in tomatoes. J. Agric. Res. 1942, 65, 0339–0346. [Google Scholar]
- Locke, S.B. Resistance to early blight and septoria leaf spot in the genus Lycopersicon. Phytopathology 1949, 39, 829–836. [Google Scholar]
- Poysa, V.; Tu, J.C. Response of cultivars and breeding lines of Lycopersicon spp., to Alternaria solani. Can. Plant Dis. Surv. 1996, 76, 5–8. [Google Scholar]
- Kalloo, G.; Banarjee, M.K. Early blight resistance in Lycopersicon esculentum Mill. transferred from L. pimpinnellifolium (L.) and L. hirsutum f. glabratum Mull. Gartenbauwissenschaft 1993, 58, 238–240. [Google Scholar]
- Reynard, G.B.; Andrus, C.F. Inheritance of resistance to the collar-rot phase of Alternaria solani on tomato. Phytopathology 1945, 35, 25–36. [Google Scholar]
- Maiero, M.; Ng, T.J.; Barksdale, T.H. Genetic resistance to early blight in tomato breeding lines. Hortscience 1990, 25, 344–346. [Google Scholar]
- Thirthamallappa; Lohithaswa, H.C. Genetics of resistance to early blight (Alternaria solani Sorauer) in tomato (Lycopersicon esculentum L.). Euphytica 2000, 113, 187–193. [Google Scholar]
- Maiero, M.; Ng, T.J.; Barksdale, T.H. Inheritance of collar rot resistance in the tomato breeding lines C1943 and NC EBR-2. Phytopathology 1990, 80, 1365–1368. [Google Scholar] [CrossRef]
- Foolad, M.R.; Lin, G.Y. Heritability of early blight resistance in a Lycopersicon esculentum × Lycopersicon hirsutum cross estimated by correlation between parent and progeny. Plant Breed. 2001, 120, 173–177. [Google Scholar] [CrossRef]
- Ashrafi, H.; Foolad, M.R. Characterization of early blight resistance in a recombinant inbred line population of tomato: II. Identification of QTLs and their co-localization with candidate resistance genes. Adv. Stud. Biol. 2015, 7, 149–168. [Google Scholar] [CrossRef]
- Ray, S.; Mondal, S.; Chowdhury, S.; Kundu, S. Differential responses of resistant and susceptible tomato varieties to inoculation with Alternaria solani. Physiol. Mol. Plant Pathol. 2015, 90, 78–88. [Google Scholar] [CrossRef]
- Kalloo, G. Genetic Improvement of Tomato; Springer-Verlag: Berlin/Heidelberg, Germany, 1991; Volume 14. [Google Scholar]
- Gardner, R.G.; Panthee, D.R. NC 1 CELBR and NC 2 CELBR: Early blight and late blight resistant fresh market tomato breeding lines. HortScience 2010, 45, 975–976. [Google Scholar]
- Barksdale, T.H. Field evaluation for tomato early blight resistance. Plant Dis. Rep. 1971, 55, 807. [Google Scholar]
- Scott, J.; Gardner, R.G. Breeding for resistance to fungal pathogens. In Genetic Improvement of Solanaceous Crops. Vol 2. Tomato; Razdan, M.K., Matton, A.K., Eds.; Science Publishers: Enfield, NH, USA, 2007; pp. 421–456. [Google Scholar]
- Ashrafi, H.; Foolad, M.R. Charaterization of early blight resistance in a recombinant inbred line population of tomato: I. Heritability and trait correlations. Adv. Stud. Biol. 2015, 7, 131–148. [Google Scholar] [CrossRef]
- Sinden, S.L.; Obrien, M.J.; Goth, R.W. Effect of potato alkaloids on growth of Alternaria solani. Am. Potato J. 1972, 49, 367. [Google Scholar]
- Johanson, A.; Thurston, H.D. The effect of cultivar maturity on the resistance of potato to early blight caused by Alternaria solani. Am. Potato J. 1990, 67, 615–623. [Google Scholar] [CrossRef]
- Martin, F.W.; Hepperly, P. Sources of resistance to early blight, Alternaria-solani, and transfer to tomato, Lycopersicon-esculentum. J. Agric. Univ. Puerto Rico 1987, 71, 85–95. [Google Scholar]
- Foolad, M.R.; Zhang, L.P.; Khan, A.A.; Nino-Liu, D.; Lin, G.Y. Identification of QTLs for early blight (Alternaria solani) resistance in tomato using backcross populations of a Lycopersicon esculentum × L-hirsutum cross. Theor. Appl. Genet. 2002, 104, 945–958. [Google Scholar] [PubMed]
- Foolad, M.R.; Panthee, D.R. Marker-assisted selection in tomato breeding. Crit. Rev. Plant Sci. 2012, 31, 93–123. [Google Scholar] [CrossRef]
- Baker, B.J. Molecular mechanisms of plant resistance to microbial disease. Mol. Biol. Cell 2000, 11, 282A. [Google Scholar]
- Zhang, L.P.; Lin, G.Y.; Nino-Liu, D.; Foolad, M.R. Mapping QTLs conferring early blight (Alternaria solani) resistance in a Lycopersicon esculentum × L. hirsutum cross by selective genotyping. Mol. Breed. 2003, 12, 3–19. [Google Scholar] [CrossRef]
- Babu, A.N.; Jogaiah, S.; Ito, S.; Nagaraj, A.K.; Tran, L.S.P. Improvement of growth, fruit weight and early blight disease protection of tomato plants by rhizosphere bacteria is correlated with their beneficial traits and induced biosynthesis of antioxidant peroxidase and polyphenol oxidase. Plant Sci. 2015, 231, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Arshad, W.; Ihsan ul, H.; Waheed, M.T.; Mysore, K.S.; Mirza, B. Agrobacterium-mediated transformation of tomato with rolB gene results in enhancement of fruit quality and foliar resistance against fungal pathogens. PLoS ONE 2014, 9, e96979. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, D.J.; St Clair, D.A. Fine mapping of three quantitative trait loci for late blight resistance in tomato using near isogenic lines (NILs) and sub-NILs. Theor. Appl. Genet. 2004, 108, 628–638. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, J.P.; Sim, S.C.; Stoffel, K.; Van Deynze, A.; Buell, C.R.; Francis, D.M. Single Nucleotide Polymorphism Discovery in Cultivated Tomato via Sequencing by Synthesis. Plant Geneme 2012, 5, 17–29. [Google Scholar] [CrossRef]
- Yang, W.C.; Bai, X.D.; Kabelka, E.; Eaton, C.; Kamoun, S.; van der Knaap, E.; Francis, D. Discovery of single nucleotide polymorphisms in Lycopersicon esculentum by computer aided analysis of expressed sequence tags. Mol. Breed. 2004, 14, 21–34. [Google Scholar] [CrossRef]
- Labate, J.A.; Baldo, A.M. Tomato SNP discovery by EST mining and resequencing. Mol. Breed. 2005, 16, 343–349. [Google Scholar] [CrossRef]
| Source of Resistance | Resulting EB-Resistant Genotype | Type of Resistance | Reference |
|---|---|---|---|
| Solanum lycopersicum | -- | ||
| Unknown source | C1943 | [84] | |
| 68B134 | 71B2 | [57] | |
| C1943 | NC63EB | [66] | |
| C1943 | NC870 | [66] | |
| C1943 | NC EBR-2 | Quantitative | [66] |
| 71B2 | NCEBR-5, NCEBR-6 | Quantitative | [67] |
| Unknown accessions | HRC90.145, HRC90.158, HRC90.159 | [73] | |
| NC-EBR-1 | NC EBR-4, IHR1816 | Quantitative | [68,77] |
| NCEBR-1 and -2 | NCEBR-3 | [68] | |
| NCEBR-3 and -4 | Mountain Supreme | [68] | |
| NCEBR-5 and -6 | Plum Dandy | [67] | |
| NCEBR-6 | Mountain Magic | [70] | |
| PI 406758 | - | [89] | |
| Solanum habrochaites | |||
| PI127827 | - | [72] | |
| PI390514, PI390662 | - | [89] | |
| PI126445 | NC EBR-1 | [66] | |
| PI1390662 | 87B187 | [76] | |
| B6013 | H-7, H-22, H-25 | [74] | |
| Unknown accessions | HRC90.303, HRC91.279, HRC91.341 | [73] | |
| LA2100, LA2124, LA2204, PE36 | - | [73] | |
| PI126445 | NC39E | [90] | |
| PI390513 | - | [89] | |
| PI390516 | - | [89] | |
| PI390658 | - | [89] | |
| PI390660 | - | [89] | |
| LA2650 | - | [73] | |
| Solanum peruvianum | |||
| PE33 | - | [73] | |
| LA1292 | - | [73] | |
| LA1365 | - | [73] | |
| LA1910 | - | [73] | |
| LA1983 | - | [73] | |
| PI270435 | - | [73] | |
| Solanum pimpinellifolium | |||
| PI 365912, PI 390519 | - | [89] | |
| A 1921 | P-1 | [74] | |
| L4394 (IHR1939) | - | [77] | |
| Resistance to collar rot | |||
| - | Devon Surprise | [71] | |
| - | Red Cherry | [71] | |
| - | Red Pear 414 | [71] | |
| - | Red Pear 415 | [71] | |
| - | Targinnie Red | [71] | |
| - | Vetomold | [71] | |
| - | Montgomery | [71] | |
| - | Norduke | [71] | |
| - | Marglobe | [71] | |
| PI179532 | - | [75] | |
| PI127833 | - | [75] | |
| No. | QTLs | Molecular Markers Associated | LOD Scores | R2-Value (%) | Parents | Type of Population | Population Size | References |
|---|---|---|---|---|---|---|---|---|
| 1 | chr1 | TG559-TG208A | 7 | 21.9 | NC84173 × PI126445 | BC1 | 145 | [90] |
| chr2 | TG337-CT59 | 2.9 | 15.3 | (S. lycopersicum × S. habrochaites) | ||||
| chr5 | XLRR.370-CT202 | 2.6 | 8.4 | |||||
| chr8 | CD40-TG16 | 2.3 | 7.4 | |||||
| chr9 | RLRR.130-CLRR.950 | 4.2 | 14.9 | |||||
| chr9 | TG424-TG429 | 5.1 | 16.2 | |||||
| chr10 | TG241-TG403 | 6.8 | 20.2 | |||||
| chr11 | CT168-TG508 | 3.8 | 13.2 | |||||
| chr11 | TG147-A41.3 | 2.3 | 7.4 | |||||
| chr12 | CT100-TG68 | 3.1 | 10.3 | |||||
| chr12 | AN23.390-TG180 | 4.1 | 12.9 | |||||
| chr1 | TG559-TG208A | 3.6 | 11.9 | NC84173 × PI126445 | BC1S1 | 145 | [90] | |
| chr2 | TG337-CT59 | 2.8 | 15.9 | (S. lycopersicum × S. habrochaites) | ||||
| chr3 | TG411-TG214 | 2.9 | 9.1 | |||||
| chr5 | TG441-CT242 | 2.6 | 7.9 | |||||
| chr5 | XLRR.370-CT202 | 3.7 | 11.3 | |||||
| chr8 | CD40-TG176 | 3 | 10.3 | |||||
| chr8 | TG330-TG294 | 5.4 | 21.0 | |||||
| chr9 | RLRR.130-CLRR.950 | 8.2 | 25.9 | |||||
| chr9 | TG424-TG429 | 3.7 | 16.2 | |||||
| chr10 | TG241-TG403 | 5.6 | 16.3 | |||||
| chr11 | TG508-TG651 | 3.8 | 11.5 | |||||
| chr11 | CT55-CD17 | 3 | 9.9 | |||||
| chr12 | CT100-TG68 | 2.5 | 8.3 | |||||
| 2 | qR_S (difference between resistant and susceptible allele frequency) | NC84173 × PI126445 | BC1 | 820 | [93] | |||
| chr3 | TG66, TG621 | ≤−0.22 | (selective genotyping) | |||||
| chr4 | TG652, PK34-340 | ≤0.28 | ||||||
| chr5 | XLRR-370, RLRR-220, S23-300, PK12-340, CT202, TG318, CT172, CT118A, TG351, CT80A, TG185 | ≤0.32 | ||||||
| chr6 | AN23-410, CT825 | ≤0.23 | ||||||
| chr8 | TG46, TG330, TG36C, CT265, CT68, TG294 | ≤0.22 | ||||||
| chr10 | CT57, CEL79 | 0.21 | ||||||
| chr11 | TG497 | 0.19 | ||||||
| 3 | COFACTOR | RAUDPC (%) | Solentos × LA2157 | F2 and F3 | 176 | [25] | ||
| chr1 | P14M60-276P | 4.1 | 6.8 | (S. lycopersicum × S. arcanum) | ||||
| chr2 | P14M51-146E | 9 | 16.2 | |||||
| chr5 | P14M51-055P | 6.2 | 10.5 | |||||
| chr6 | P11M48-266E | 6.3 | 10.8 | |||||
| chr7 | P15M62-349P | 8.3 | 15.2 | |||||
| chr9 | P11M60-109P | 8.7 | 15.5 | |||||
| 4 | chr2, chr3, chr4, chr5, chr6, chr7, chr9, and chr12 | - | - | combined effect = 44% | NCEBR1 × LA2093 | F2, F3, F4 | - | [18] |
| Individual = 7.6% to13.4% | (S. lycopersicum × S. pimpinellifolium) | |||||||
| 5 | chr1, chr2, chr3, chr4, chr5, chr6, and chr11 | - | - | - | NCEBR1 × LA2093 | F2 | - | [18] |
| (Selective genotyping) | ||||||||
| 6 | chr2 | cLEC73K6b, cLEC73I19a, CT205 | 3.3 | 8.0 | NCEBR1 × LA2093 | RIL | 172 | [80] |
| chr2 | TG463, cTOFC19J9, CT103 | 2.9 | 5.5 | |||||
| chr5 | cLEY-18H8, cTOA24J24, cTOC2J24, cTOC20J21 | 5.5 | 15.3 | |||||
| chr6 | TG274, cLEN10H12, TG590, cLEN10H12 | 4.4 | 13.0 | |||||
| chr9 | TG343, cLED4N20, TG348, cTOE10J18 | 4 | 11.5 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adhikari, P.; Oh, Y.; Panthee, D.R. Current Status of Early Blight Resistance in Tomato: An Update. Int. J. Mol. Sci. 2017, 18, 2019. https://doi.org/10.3390/ijms18102019
Adhikari P, Oh Y, Panthee DR. Current Status of Early Blight Resistance in Tomato: An Update. International Journal of Molecular Sciences. 2017; 18(10):2019. https://doi.org/10.3390/ijms18102019
Chicago/Turabian StyleAdhikari, Pragya, Yeonyee Oh, and Dilip R. Panthee. 2017. "Current Status of Early Blight Resistance in Tomato: An Update" International Journal of Molecular Sciences 18, no. 10: 2019. https://doi.org/10.3390/ijms18102019




