Peroxisome Proliferator-Activated Receptor γ Expression Is Inversely Associated with Macroscopic Vascular Invasion in Human Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Results
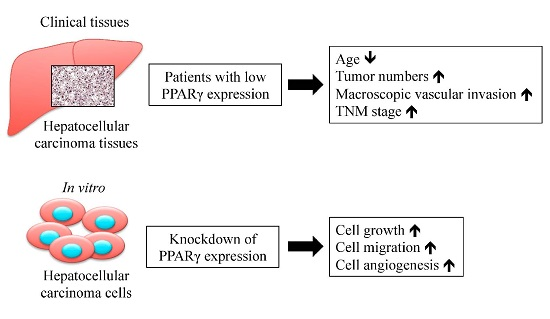
2.1. Peroxisome Proliferator-Activated Receptor γ (PPARγ) Protein Expression in Human Hepatocellular Carcinoma (HCC) Tissues and Associated Clinicopathological Characteristics
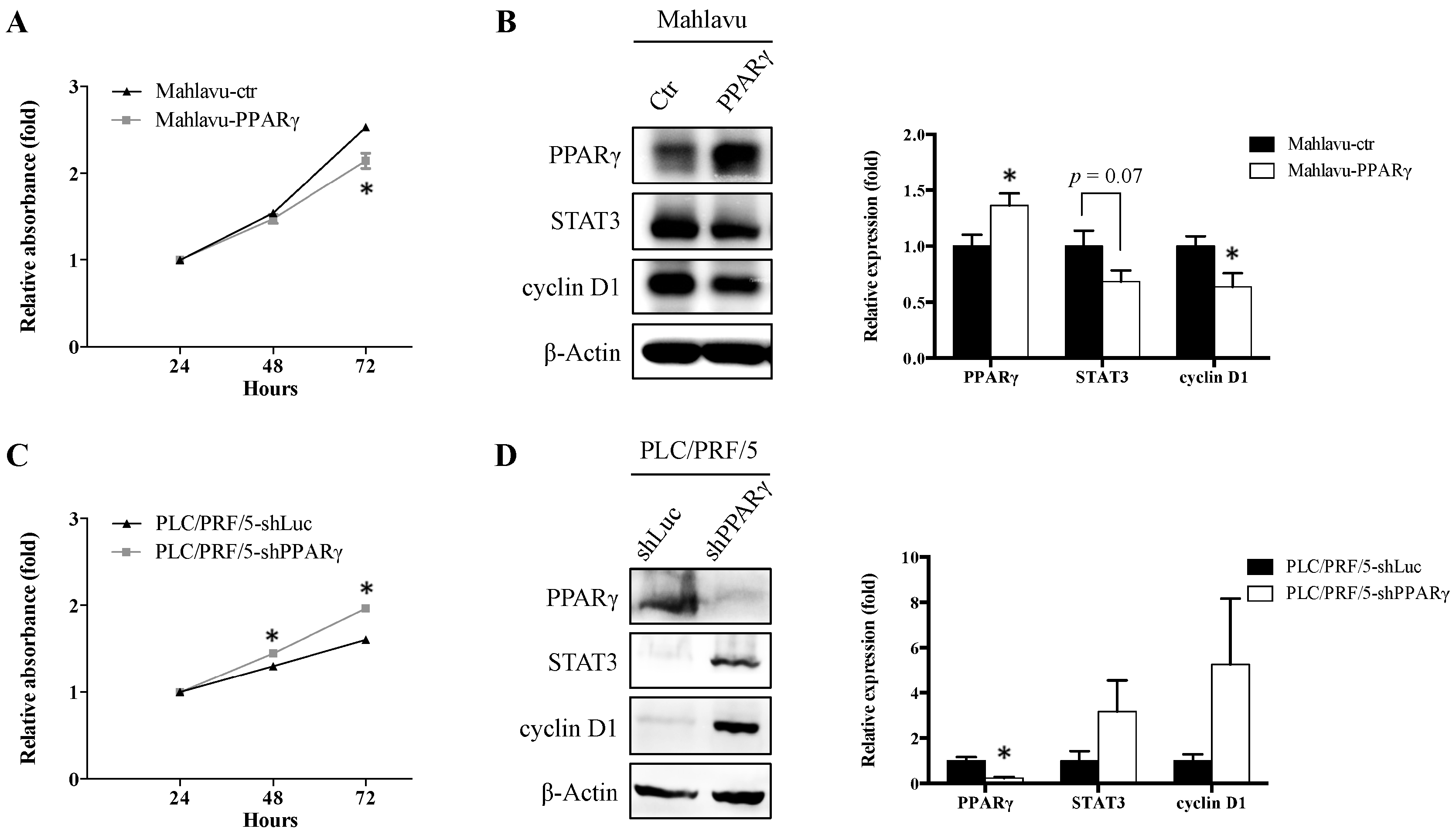
2.2. PPARγ Suppresses HCC Cell Proliferation
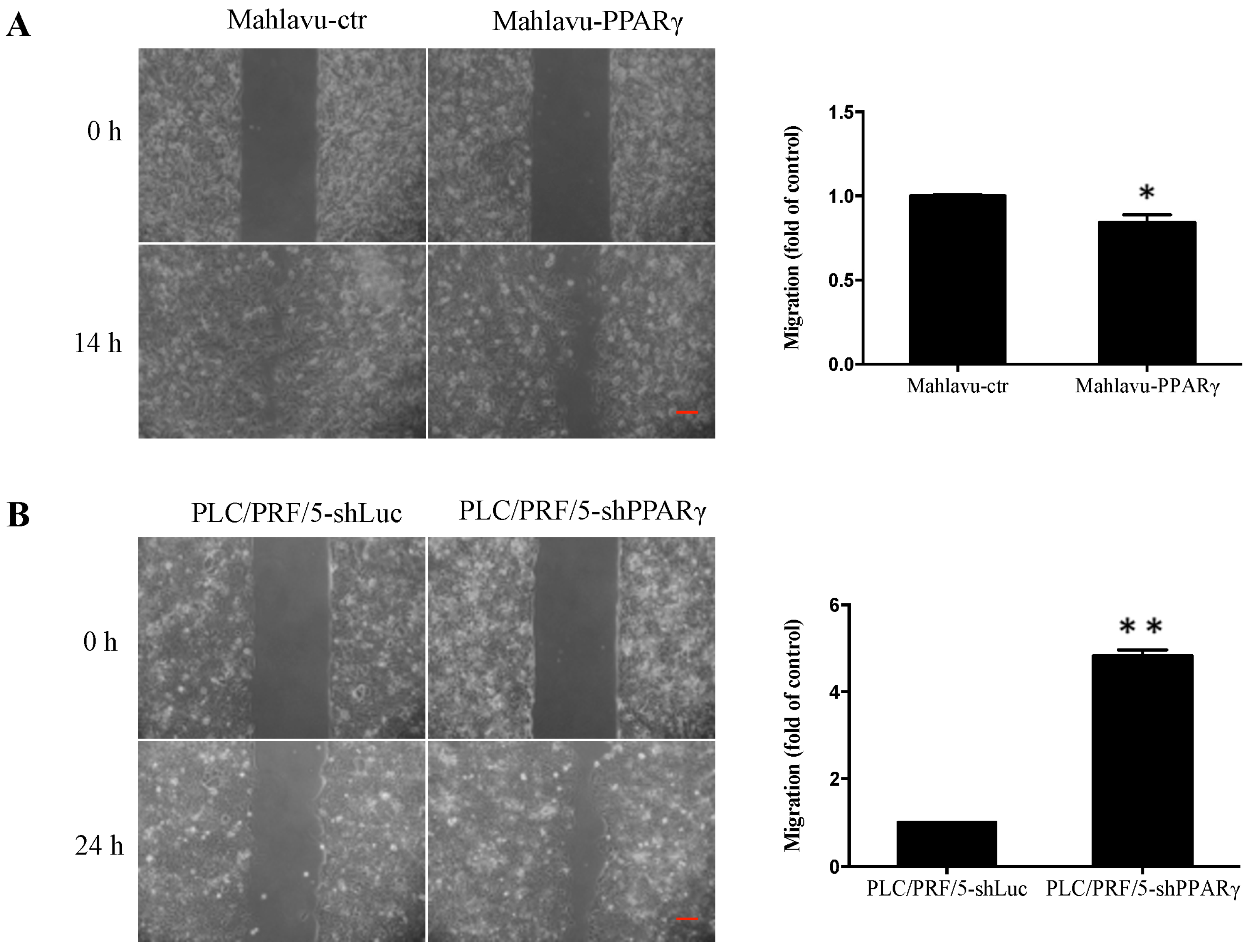
2.3. PPARγ Inhibits HCC Cell Migration
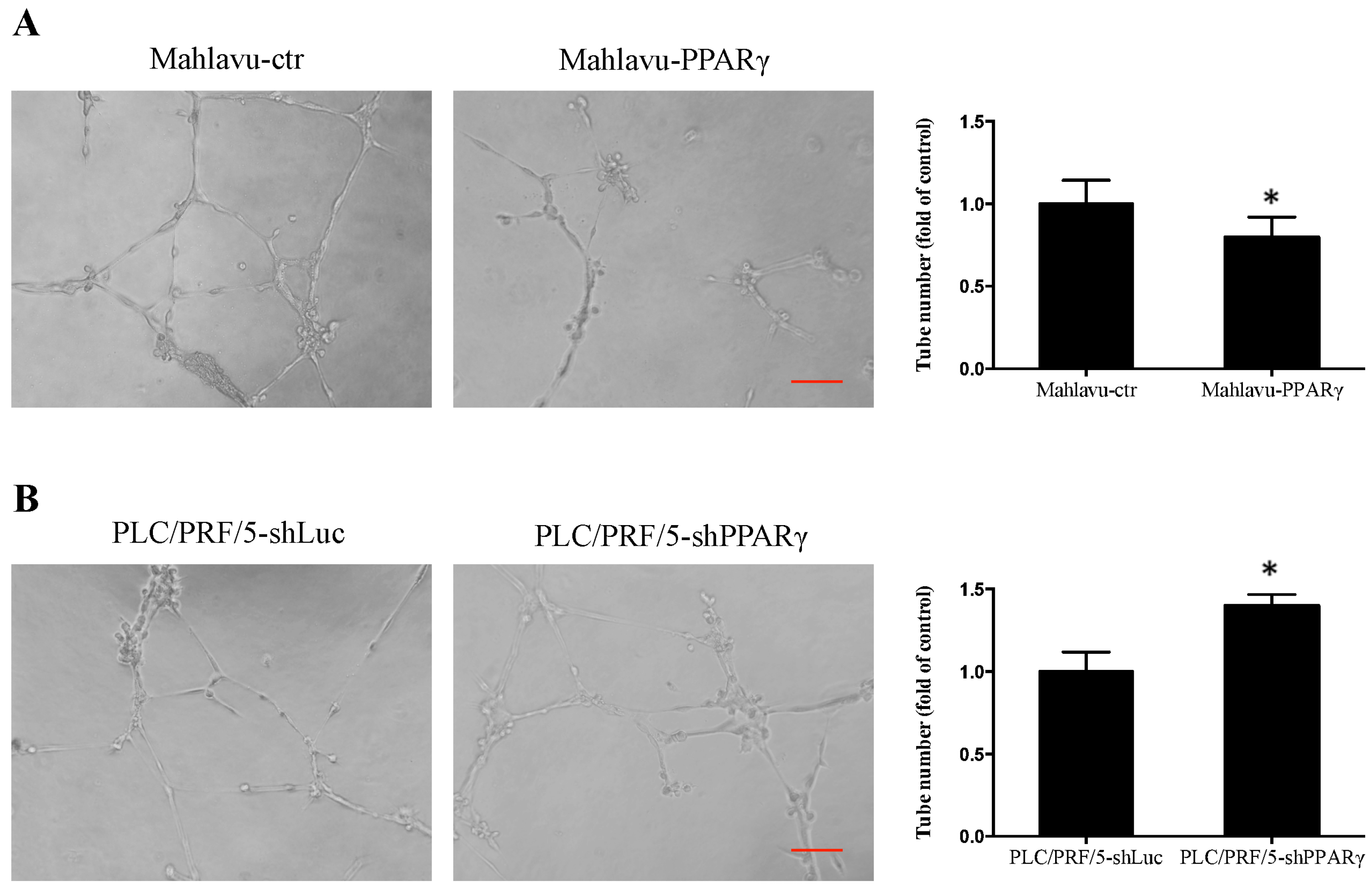
2.4. PPARγ Decreases HCC Cell Angiogenesis
3. Discussions
4. Materials and Methods
4.1. Human Tissue Specimens and Patient Information
4.2. Immunohistochemical
4.3. Cell Culture
4.4. PPARγ Overexpression and Knockdown
4.5. Cell Proliferation Measurement
4.6. Western Blot Analyses
4.7. Wound Healing Assay
4.8. Matrigel Tube Formation Assay
4.9. Statistical Analyses
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012, 142, 1264–1273. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.Y. Hepatocellular carcinoma—Cause, treatment and metastasis. World J. Gastroenterol. 2001, 7, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Desvergne, B.; Wahli, W. Peroxisome proliferator-activated receptors: Nuclear control of metabolism. Endocr. Rev. 1999, 20, 649–688. [Google Scholar] [CrossRef] [PubMed]
- Forman, B.M.; Tontonoz, P.; Chen, J.; Brun, R.P.; Spiegelman, B.M.; Evans, R.M. 15-Deoxy-∆12,14-prostaglandin J2 is a ligand for the adipocyte determination factor PPARγ. Cell 1995, 83, 803–812. [Google Scholar] [CrossRef]
- Murphy, G.J.; Holder, J.C. PPAR-γ agonists: Therapeutic role in diabetes, inflammation and cancer. Trends Pharmacol. Sci. 2000, 21, 469–474. [Google Scholar] [CrossRef]
- Cao, L.Q.; Shao, Z.L.; Liang, H.H.; Zhang, D.W.; Yang, X.W.; Jiang, X.F.; Xue, P. Activation of peroxisome proliferator-activated receptor-γ (PPARγ) inhibits hepatoma cell growth via downregulation of SEPT2 expression. Cancer Lett. 2015, 359, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Su, Y.; Yin, P.H.; Lee, H.C.; Chi, C.W. PPARγ/PGC-1α pathway in E-cadherin expression and motility of HepG2 cells. Anticancer Res. 2009, 29, 5057–5063. [Google Scholar] [PubMed]
- Gu, J.J.; Zhang, J.H.; Chen, H.J.; Wang, S.S. MicroRNA-130b promotes cell proliferation and invasion by inhibiting peroxisome proliferator-activated receptor-γ in human glioma cells. Int. J. Mol. Med. 2016, 37, 1587–1593. [Google Scholar] [CrossRef] [PubMed]
- Ohta, K.; Endo, T.; Haraguchi, K.; Hershman, J.M.; Onaya, T. Ligands for peroxisome proliferator-activated receptor γ inhibit growth and induce apoptosis of human papillary thyroid carcinoma cells. J. Clin. Endocrinol. Metab. 2001, 86, 2170–2177. [Google Scholar] [PubMed]
- Shen, B.; Chu, E.S.; Zhao, G.; Man, K.; Wu, C.W.; Cheng, J.T.; Li, G.; Nie, Y.; Lo, C.M.; Teoh, N.; et al. PPARγ inhibits hepatocellular carcinoma metastases in vitro and in mice. Br. J. Cancer 2012, 106, 1486–1494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Shen, B.; Chu, E.S.; Teoh, N.; Cheung, K.F.; Wu, C.W.; Wang, S.; Lam, C.N.; Feng, H.; Zhao, J.; et al. Inhibitory role of peroxisome proliferator-activated receptor γ in hepatocarcinogenesis in mice and in vitro. Hepatology 2010, 51, 2008–2019. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Wei, Y.; Zhang, Y.; Zhang, M.; Lu, Y.; Shen, P. Peroxisome proliferator-activated receptor-γ activation inhibits hepatocellular carcinoma cell invasion by upregulating plasminogen activator inhibitor-1. Cancer Sci. 2013, 104, 672–680. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Gao, Y.; Jia, Z.; Mishra, L.; Guo, K.; Li, Z.; Le, X.; Wei, D.; Huang, S.; Xie, K. Dysregulated kruppel-like factor 4 and vitamin D receptor signaling contribute to progression of hepatocellular carcinoma. Gastroenterology 2012, 143, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Sung, M.T.; Hsu, H.T.; Lee, C.C.; Lee, H.C.; Kuo, Y.J.; Hua, K.; Hsia, C.Y.; Chi, C.W. Kruppel-like factor 4 modulates the migration and invasion of hepatoma cells by suppressing TIMP-1 and TIMP-2. Oncol. Rep. 2015, 34, 439–446. [Google Scholar] [PubMed]
- Lin, Z.S.; Chu, H.C.; Yen, Y.C.; Lewis, B.C.; Chen, Y.W. Kruppel-like factor 4, a tumor suppressor in hepatocellular carcinoma cells reverts epithelial mesenchymal transition by suppressing slug expression. PLoS ONE 2012, 7, e43593. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhou, Q.; He, H.; Zhao, Y.; Liu, Z. Peroxisome proliferator-activated receptor γ agonists induce cell cycle arrest through transcriptional regulation of kruppel-like factor 4 (KLF4). J. Biol. Chem. 2013, 288, 4076–4084. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, K.L.; Wada, K.; Takahashi, H.; Matsuhashi, N.; Ohnishi, S.; Wolfe, M.M.; Turner, J.R.; Nakajima, A.; Borkan, S.C.; Saubermann, L.J. Peroxisome proliferator-activated receptor γ inhibition prevents adhesion to the extracellular matrix and induces anoikis in hepatocellular carcinoma cells. Cancer Res. 2005, 65, 2251–2259. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.M.; Velmurugan, B.K.; Yeh, Y.L.; Tu, C.C.; Ho, T.J.; Lai, T.Y.; Tsai, C.H.; Tsai, F.J.; Tsai, C.H.; Huang, C.Y. Activation of estrogen receptors with E2 downregulates peroxisome proliferator-activated receptor γ in hepatocellular carcinoma. Oncol. Rep. 2013, 30, 3027–3031. [Google Scholar] [PubMed]
- Yu, J.; Qiao, L.; Zimmermann, L.; Ebert, M.P.; Zhang, H.; Lin, W.; Rocken, C.; Malfertheiner, P.; Farrell, G.C. Troglitazone inhibits tumor growth in hepatocellular carcinoma in vitro andin vivo. Hepatology 2006, 43, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Vitale, G.; Zappavigna, S.; Marra, M.; Dicitore, A.; Meschini, S.; Condello, M.; Arancia, G.; Castiglioni, S.; Maroni, P.; Bendinelli, P.; et al. The PPAR-γ agonist troglitazone antagonizes survival pathways induced by stat-3 in recombinant interferon-β treated pancreatic cancer cells. Biotechnol. Adv. 2012, 30, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhao, L.H.; Huang, B.; Wang, R.Y.; Yuan, S.X.; Tao, Q.F.; Xu, Y.; Sun, H.Y.; Lin, C.; Zhou, W.P. Pioglitazone, a PPARγ agonist, inhibits growth and invasion of human hepatocellular carcinoma via blockade of the rage signaling. Mol. Carcinog. 2015, 54, 1584–1595. [Google Scholar] [CrossRef] [PubMed]
- Faber, W.; Stockmann, M.; Kruschke, J.E.; Denecke, T.; Bahra, M.; Seehofer, D. Implication of microscopic and macroscopic vascular invasion for liver resection in patients with hepatocellular carcinoma. Dig. Surg. 2014, 31, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Roayaie, S.; Frischer, J.S.; Emre, S.H.; Fishbein, T.M.; Sheiner, P.A.; Sung, M.; Miller, C.M.; Schwartz, M.E. Long-term results with multimodal adjuvant therapy and liver transplantation for the treatment of hepatocellular carcinomas larger than 5 centimeters. Ann. Surg. 2002, 235, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.W.; Farrell, G.C.; Yu, J. Functional role of peroxisome-proliferator-activated receptor γ in hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2012, 27, 1665–1669. [Google Scholar] [CrossRef] [PubMed]
- Panigrahy, D.; Huang, S.; Kieran, M.W.; Kaipainen, A. PPARγ as a therapeutic target for tumor angiogenesis and metastasis. Cancer Biol. Ther. 2005, 4, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Panigrahy, D.; Singer, S.; Shen, L.Q.; Butterfield, C.E.; Freedman, D.A.; Chen, E.J.; Moses, M.A.; Kilroy, S.; Duensing, S.; Fletcher, C.; et al. PPARγ ligands inhibit primary tumor growth and metastasis by inhibiting angiogenesis. J. Clin. Investig. 2002, 110, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Peeters, L.L.; Vigne, J.L.; Tee, M.K.; Zhao, D.; Waite, L.L.; Taylor, R.N. PPAR γ represses VEGF expression in human endometrial cells: Implications for uterine angiogenesis. Angiogenesis 2005, 8, 373–379. [Google Scholar] [CrossRef] [PubMed]
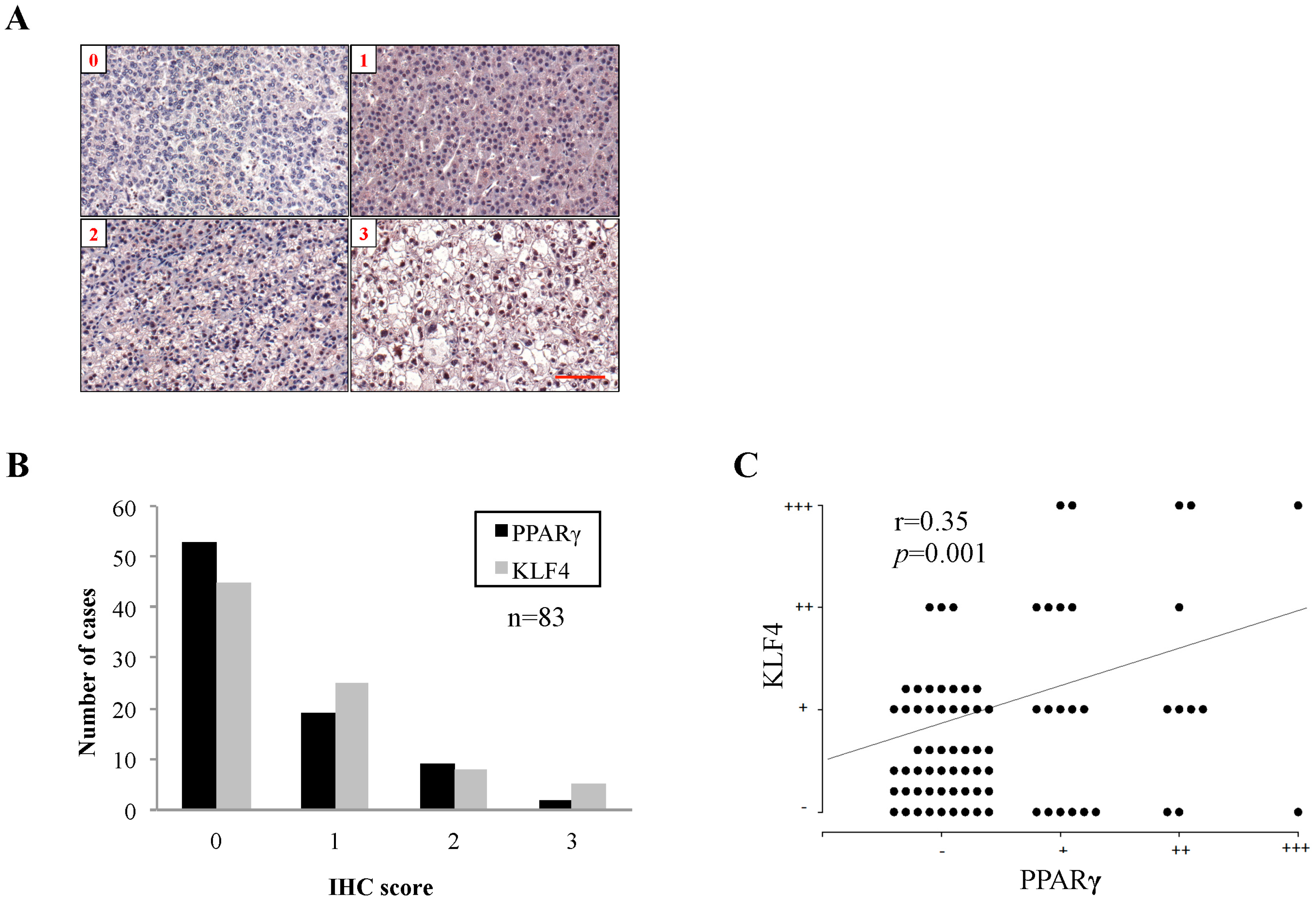
| Characteristic | PPARγ Expression (Percentage) | p Value | |
|---|---|---|---|
| Low (=0) n = 53 | High (>0) n = 30 | ||
| Age | |||
| ≤65 | 39 | 13 | 0.006 * |
| >65 | 14 | 17 | |
| Sex | |||
| Male | 39 | 21 | 0.726 |
| Female | 14 | 9 | |
| Tumor size | |||
| <3 cm | 8 | 9 | 0.106 |
| >3 cm | 45 | 21 | |
| HBsAg ∆ | |||
| (−) | 15 | 14 | 0.053 |
| (+) | 38 | 14 | |
| Anti-HCV ∆ | |||
| (−) | 44 | 22 | 0.148 |
| (+) | 7 | 8 | |
| Cell differentiation | |||
| Well differentiated | 3 | 3 | 0.63 |
| Moderately differentiated | 32 | 19 | |
| Poorly differentiated | 18 | 8 | |
| Tumor number ∆ | |||
| Single | 33 | 25 | 0.038 * |
| Multiple | 18 | 4 | |
| Liver cirrhosis | |||
| No | 42 | 21 | 0.344 |
| Yes | 11 | 9 | |
| Chronic hepatitis ∆ | |||
| No | 6 | 7 | 0.227 |
| Yes | 41 | 23 | |
| Fibrosis ∆ | |||
| No | 29 | 17 | 0.293 |
| Yes | 12 | 12 | |
| MVI | |||
| No | 36 | 28 | 0.008 * |
| Yes | 17 | 2 | |
| Bile duct invasion | |||
| No | 49 | 29 | 0.649 |
| Yes | 4 | 1 | |
| AFP ∆ | |||
| <20 | 19 | 16 | 0.139 |
| >20 | 33 | 14 | |
| TNM stage | |||
| I + II | 30 | 25 | 0.013 * |
| III | 23 | 5 | |
| DFS | |||
| Event/all | 34/53 | 18/30 | 0.967 |
| 5-year survival | 34.6% | 38% | |
| OS | |||
| Event/all | 25/53 | 15/30 | 0.349 |
| 5-year survival | 55.3% | 46.6% | |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, H.-T.; Sung, M.-T.; Lee, C.-C.; Kuo, Y.-J.; Chi, C.-W.; Lee, H.-C.; Hsia, C.-Y. Peroxisome Proliferator-Activated Receptor γ Expression Is Inversely Associated with Macroscopic Vascular Invasion in Human Hepatocellular Carcinoma. Int. J. Mol. Sci. 2016, 17, 1226. https://doi.org/10.3390/ijms17081226
Hsu H-T, Sung M-T, Lee C-C, Kuo Y-J, Chi C-W, Lee H-C, Hsia C-Y. Peroxisome Proliferator-Activated Receptor γ Expression Is Inversely Associated with Macroscopic Vascular Invasion in Human Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2016; 17(8):1226. https://doi.org/10.3390/ijms17081226
Chicago/Turabian StyleHsu, Hui-Tzu, Ming-Ta Sung, Chih-Chun Lee, Yin-Ju Kuo, Chin-Wen Chi, Hsin-Chen Lee, and Cheng-Yuan Hsia. 2016. "Peroxisome Proliferator-Activated Receptor γ Expression Is Inversely Associated with Macroscopic Vascular Invasion in Human Hepatocellular Carcinoma" International Journal of Molecular Sciences 17, no. 8: 1226. https://doi.org/10.3390/ijms17081226