1. Introduction
The gradually increasing prevalence of obesity has caused adipose tissue to be a focus. Adipose tissue, which has been divided into two main types, white adipose tissue (WAT) and brown adipose tissue (BAT), is a complex and highly active metabolic organ [
1]. WAT, best known for its role in the storage of energy, is distributed mainly in visceral and subcutaneous sites. BAT is viewed as a key site of nonshivering thermogenesis and has been considered an attractive target for treating obesity and obesity-associated diseases [
2]. Most BAT depots are located in the interscapular and perirenal regions in rodents and large mammals [
3]. Recently, a third class of adipocytes was discovered in WAT, especially in inguinal WAT, and the cells were termed “beige” or “brown-like” adipocytes [
3]. Interestingly, even though beige adipocytes exist in WAT, beige and brown adipocytes have similar morphology, in the form of multilocular lipid droplets, and a common ability to carry out thermogenesis [
2]. In fact, increasing evidence suggested that WAT browning and BAT activation may be promising strategies for treating obesity [
4,
5,
6]. However, there is still relatively little knowledge of the relations between WAT and BAT, especially at the molecular level. In order to study depot-related differences in mRNA expression in adipose tissue, a sensitive and specific technique, such as quantitative real-time polymerase chain reaction (qRT-PCR) is required [
7].
Actually, qRT-PCR is a valuable tool for gene expression analysis and is used in many clinical and scientific research fields [
8]. However, many studies have demonstrated that the results of qRT-PCR are affected by experimental operation, original sample amount, RNA integrity, cDNA quality and the efficiency of reverse transcription [
9,
10]. Thus, choosing a stable reference gene for data normalization is the most primary step of a qRT-PCR assay [
11]. However, no universal reference gene has been confirmed, and reference genes are regulated by the certain experimental conditions [
12]. Therefore, the stability of frequently-used reference genes is relative. From a survey of different studies of adipose tissue, qRT-PCR data were normalized to different reference genes, such as Glyceraldehyde-3-phosphate dehydrogenase (
GAPDH) [
13],
β-actin [
14],
18S [
15] and acidic ribosomal phosphoprotein P0 (
36B4) [
16]. However, no research has studied the stability of frequently-used reference genes in different types of rat adipose tissue.
Since 2002, several mathematical algorithms, such as geNorm [
17], NormFinder [
10], and BestKeeper [
18], have been applied to evaluate the expression stability of reference genes. GeNorm evaluates the most stable reference genes for normalization determined by the M value, which is calculated from the arithmetic mean of pairwise variations of each gene. NormFinder validates the expression stability of each reference gene and consider both intra- and intergroup variations for normalization. BestKeeper calculates the expression variability of reference genes based on the standard deviation (SD), coefficient of variance (CV) and coefficient of correlation. In the comprehensive ranking, the top ranked genes are recommended as reference genes for data normalization in similar experimental systems.
In 2009, the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines proposed that selecting multiple suitable reference genes for reliable normalization in qRT-PCR is necessary [
19]. Therefore, in this study, we used geNorm, NormFinder and BestKeeper to identify four commonly used reference genes in epididymal WAT (eWAT), inguinal beige adipose tissues (iBeAT) and brown adipose tissue (BAT) from obese and lean rats, and to identify the two most suitable reference genes for data normalization. Furthermore, we used two target genes to confirm how the different reference genes affect the final results.
3. Discussion
In molecular biological research, gene expression analysis is one of the most frequently-used strategies in the field of gene study involved in signaling and metabolic pathways. It is of vital significance to select an appropriate reference gene for the normalization of gene expression. It is generally known that
GAPDH and
β-actin are the two most commonly-used reference genes. However, some researchers have identified that
GAPDH and
β-actin showed a significant variation in different cells, tissues and experimental conditions [
24,
25,
26]. Furthermore, Catalan and his colleagues showed that
GAPDH is a less appropriate reference gene in human omental and subcutaneous adipose tissue from obesity and type 2 diabetes patients, because of the variational expression [
27]. Therefore, we identified the expression stability of four reference genes (
GAPDH,
β-actin,
36B4 and
18S) in three types of rat adipose tissue (eWAT, iBeAT and BAT) by using different software: geNorm, NormFinder and BestKeeper. In addition, in accordance with the MIQE guidelines, the two most suitable reference genes were recommended in each type of adipose tissue.
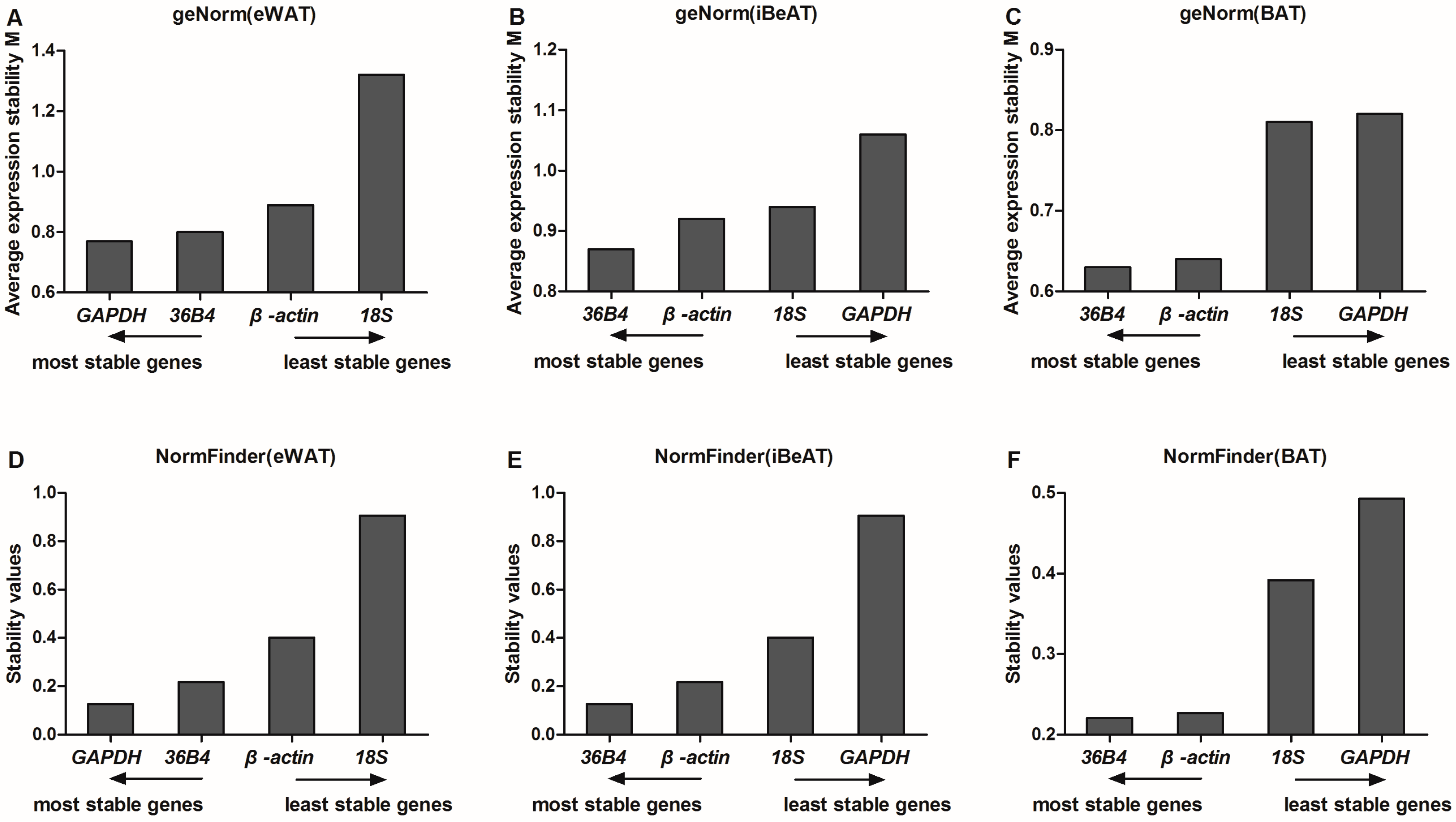
GeNorm software is one of the most commonly-used algorithms applied to evaluate reference gene expression stability via a pairwise comparison between one reference gene and each other reference gene from each sample [
17]. In our study,
GAPDH and
36B4 appeared to be the two most stable reference genes in eWAT among lean and obese rats. However,
GAPDH was the least stable reference gene in both iBeAT and BAT. Therefore,
GAPDH is not a universal reference gene in adipose tissue. This finding is consistent with a previous study by Akamine et al
., who verified that
GAPDH was not suitable as a universal reference gene in adipose tissue because of its inconsistent expression levels between WAT and BAT [
28]. In our study, however, we found that
36B4 and
β-actin had highest expression stability in both iBeAT and BAT. It is suggested that
36B4 is the universal reference gene in different types of adipose tissue.
Unlike geNorm, the NormFinder procedure directly and robustly estimates gene expression stability and ranks reference genes depend on the variation within the intra- and the inter-group. As Andersen mentioned in his report, the NormFinder procedure focuses on differences between sample subgroups, and the result is less affected by the correlated expression of the reference genes [
10]. In our study, the results obtained from NormFinder analysis were quite consistent with the geNorm analysis.
GAPDH is the most stable reference gene, followed by
36B4 in eWAT.
36B4 and
β-actin were the most stable reference genes in both iBeAT and BAT.
BestKeeper is another traditional software that is used to assess the stability of gene expression. In the BestKeeper procedure, evaluating the stability of gene expression was co-determined by three essential criteria, which are the SD, CV and the coefficient of correlation [
18]. In our study,
36B4 was the most stable gene in eWAT based on the CV value, while
GAPDH and
β-actin were ranked as the second and third, respectively; these results were different from the geNorm and NormFinder analysis. In terms of the coefficient of correlation, however,
GAPDH was the most stable gene in eWAT. In addition, this result completely supports the geNorm and NormFinder analysis. In iBeAT,
36B4 had a highest expression stability based on both CV value and coefficient of correlation. This result was consistent with the geNorm and NormFinder analysis. In BAT, a difference was also observed based these two criteria. When ordered by CV value and coefficient of correlation,
36B4 and
β-actin were the most stable genes, respectively.
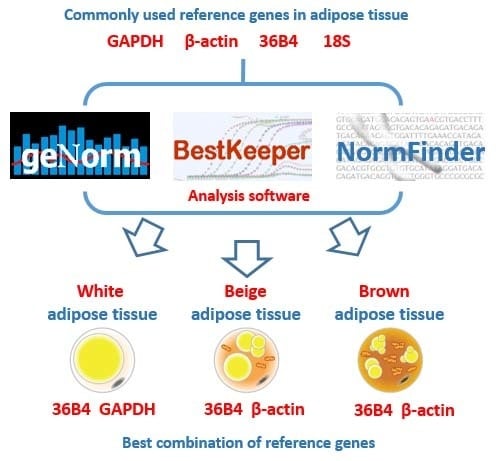
To evaluate the different results from the three algorithms systematically, a comprehensive ranking of each reference gene was calculated. Results show that
GAPDH was the most stable reference gene in eWAT, but it had the highest variability in both iBeAT and BAT. In addition,
36B4 was a highly stable gene in each type of adipose tissue, even though the results from the geNorm and NormFinder analyses had a tiny disparity with
GAPDH in eWAT. For a realistic study of obesity, more than one type of adipose tissue should be considered. Based on new research, “browning” of WAT will become a new strategy for obesity treatment [
4,
29]. Therefore, considering the variety of experimental subjects and practicability, a relatively universal reference gene among each type of adipose tissue should be used. According to our study,
36B4 is the universal reference gene in adipose tissue. In accordance with the MIQE guidelines, however, the use of multiple reference genes for normalization in qRT-PCR were proposed
. We recommend
36B4 and
GAPDH as the best combination of reference genes for eWAT, and
36B4 and
β-actin as the most suitable reference genes for both iBeAT and BAT. To further validate our assumptions in an actual experiment, we used either the combination of most or the least stable reference gene to normalize same target genes (e.g.,
leptin and
UCP-1). As shown in
Figure 4, expected results and statistical significance were obtained by normalizing with the two most stable reference genes. This result totally confirmed our assumptions.
4. Experimental Section
4.1. Animals
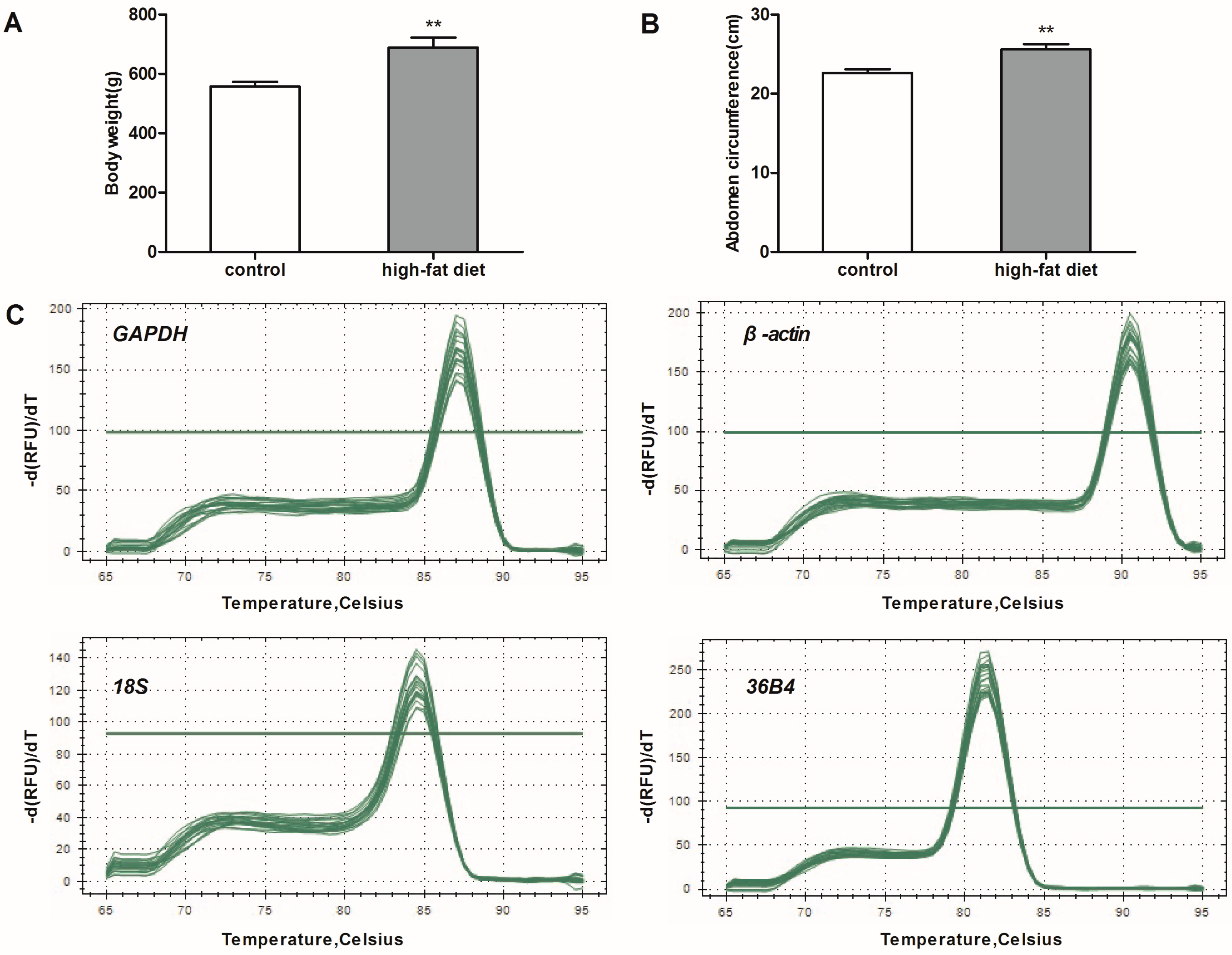
Six-week-old male Wistar rats weighing 150–200 g were provied by the Experimental Center of Hubei Medical Scientific Academy (No. 2008-0005, Wuhan, Hubei, China). Rats were maintained with 12 h light-dark cycles at 22–24 °C room temperature. Rats were randomly assigned to two groups: a high-fat diet group (n = 6) and a control group (n = 6). The rats in the control group were fed a standard chow diet and water ad libitum. Meanwhile, rats in high-fat diet group were fed a high-fat diet (45% calories from fat, D12451) for 20 weeks. The body weight of each rat was monitored weekly until 26 weeks of age. At the end of the experiment, rats fasted for 12 h and were then placed in a separate quiet room for anesthesia. After the disappearance of the righting reflex, the abdominal circumference of animal was measured. Rats were killed by rapid decapitation. eWAT, iBeAT and BAT were collected. Adipose tissues were immediately frozen in liquid nitrogen and later stored at −80 °C for subsequent experiments.
4.2. RNA Isolation, Quantification and cDNA Synthesis
The total RNA of eWAT, iBeAT and BAT were extracted using the Trizol instructed by the manufacturer’s protocol. The oil was removed by repeated centrifugation. The purity of the total RNA was performed on a NanoDrop 2000c spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Only RNA samples with a 260/280 ratio between 1.8 and 2.1 were used in subsequent analyses. cDNA was synthesized from 1 μg of total RNA from eWAT, iBeAT and BAT in control and high-fat diet rats using the PrimeScript RT-PCR kit (Takara Biotech, Kusatsu, Japan) instructed by the manufacturer’s recommendations. After synthesis, the template for qRT-PCR was prepared for further studies by diluting the obtained cDNA tenfold with ultrapure water.
4.3. qRT-PCR
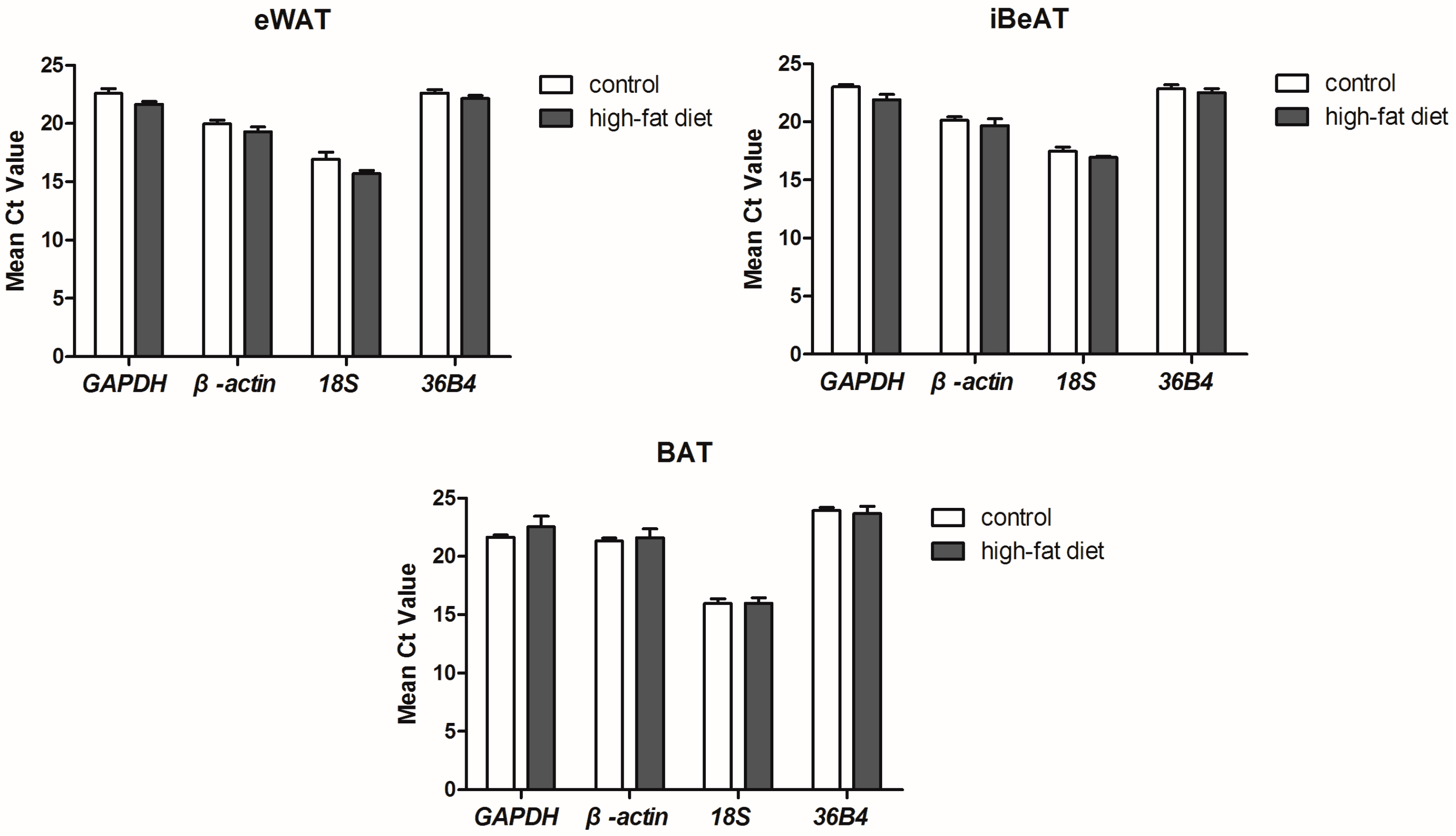
qRT-PCR was conducted on a StepOnePlus™ real-time PCR system (Applied BioSystems, Foster City, CA, USA). Each reaction contained 1 μL of cDNA template and 9 μL of the reaction mixture, which were used in the SYBR Green Core Reagent kit (Applied Biosystems, Foster City, CA, USA). Relative mRNA expression levels were calculated by the ΔΔ
Ct method. The cycling condition is as follows: pre-denaturation, 95 °C for 30 s; denaturation, 95 °C for 10 s; annealing, 60 °C for 30 s; 40 cycles. Then, a melting curve was generated by increasing the temperature from 65 to 95 °C with continuous collection of the SYBR Green fluorescence signal. All primer sequences and relevant parameters about the genes are showed in
Table 7.
4.4. Reference Gene Expression Stability Analysis
The initial qRT-PCR data were obtained from the StepOnePlus™ software (Applied BioSystems, Foster City, CA, USA) and put into an Excel datasheet. Then, statistical tools such as GeNorm [
17], NormFinder [
10] and BestKeeper Version [
18] were used to evaluate the stability of gene expression.
4.5. Statistical Analysis
Data were presented as the mean ± SEM. Graphics were performed using Prism (GraphPad Software, Version 5.0, La Jolla, CA, USA). All results were evaluated using an unpaired Student’s t-test, and differences with p < 0.05 were regarded as statistically significant.