Cloning, Expression and 3D Structure Prediction of Chitinase from Chitinolyticbacter meiyuanensis SYBC-H1
Abstract
:1. Introduction
2. Results and Discussion
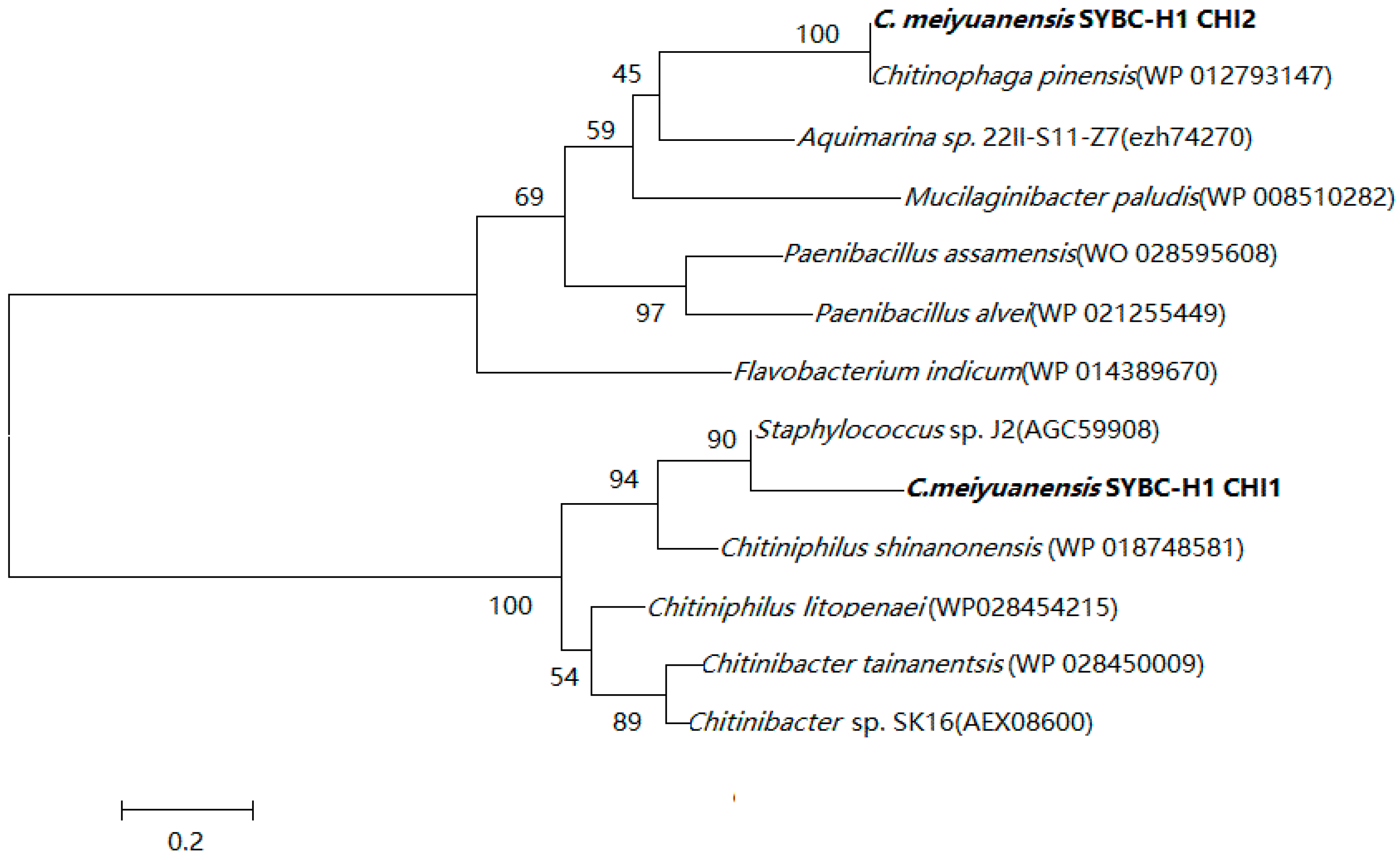
2.1. Sequence Alignment with Chitinase
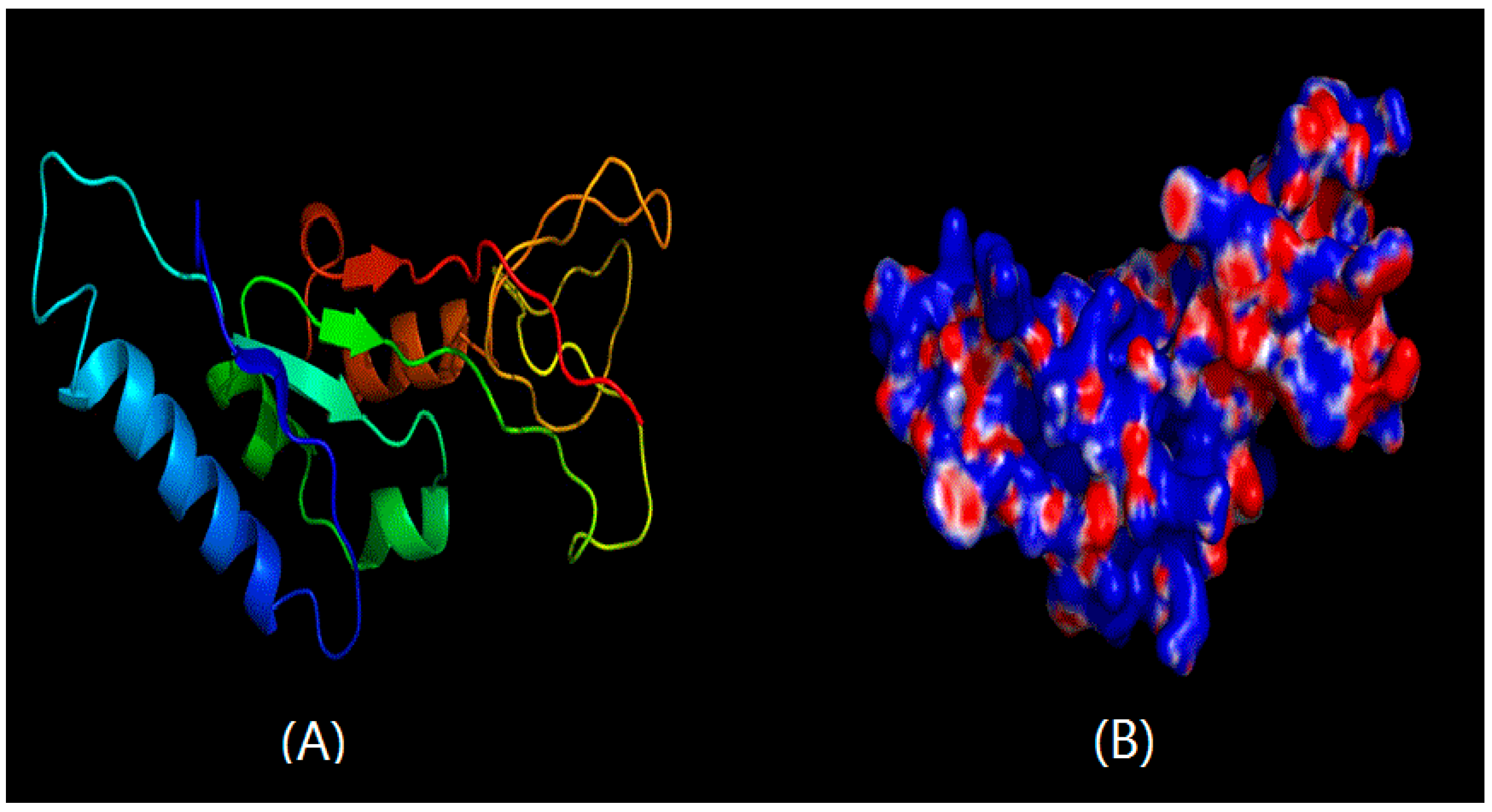
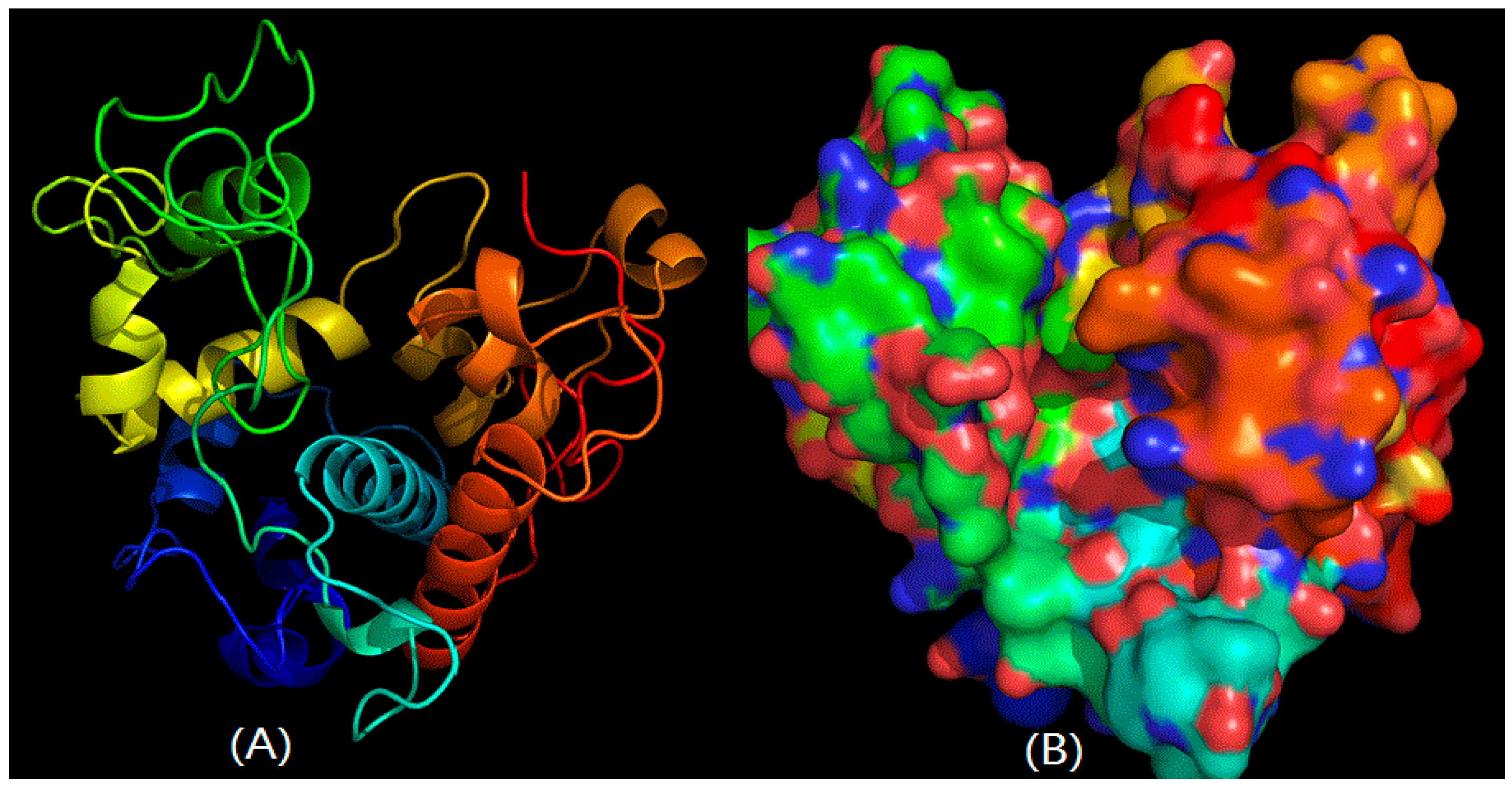
2.2. Structural Prediction of CHI Chitinase
2.3. Expression, Purification of CHI2 Protein in E. coli
2.4. Characterization of Purified Chitinase
2.5. Effects of Temperature and pH on the CHI2 Chitinase
2.6. Effects of Metal Ions and Chemicals on the CHI2 Chitinase Activity
2.7. Kinetic Study and Chitin Hydrolysis
3. Materials and Methods
3.1. Microorganism and Cultivation
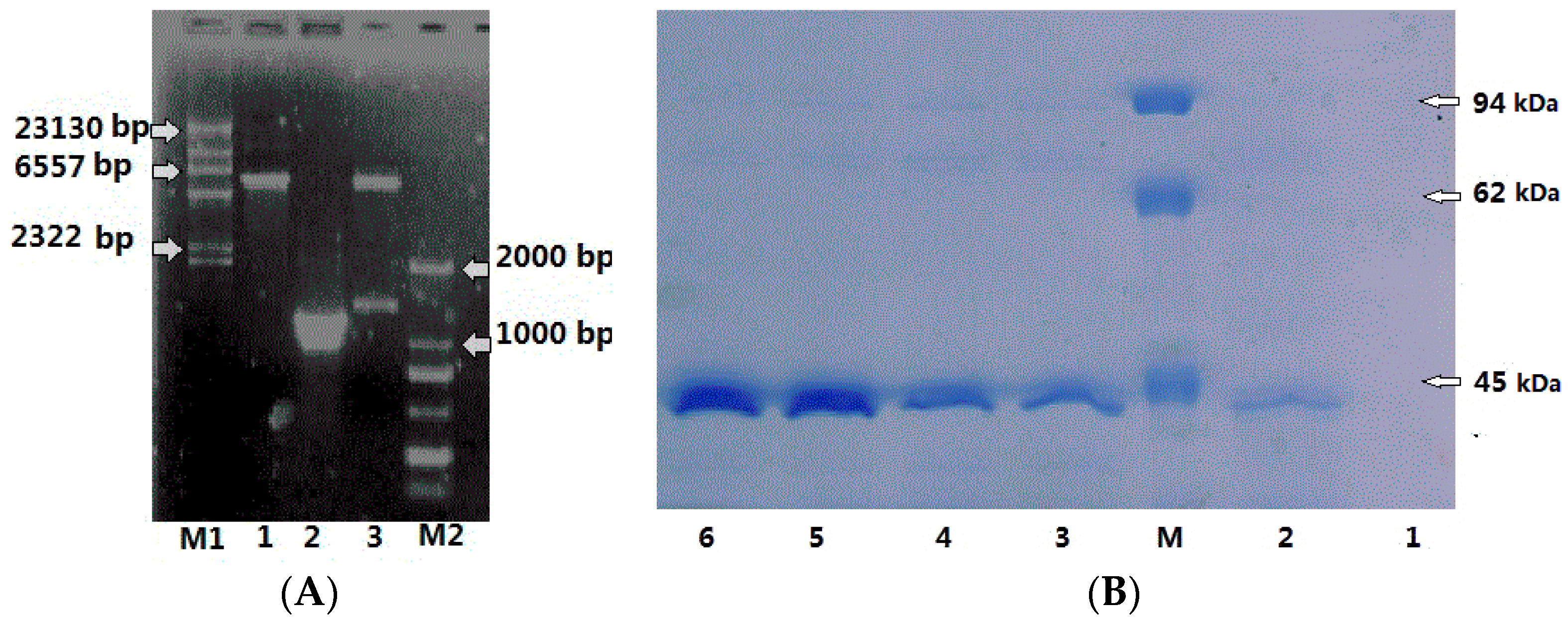
3.2. Cloning of CHI1 and CHI2 Gene
3.3. Sequence Analysis and Structure Prediction
3.4. Kinetic Methods
3.5. Purification of CHI Chitinase
3.6. Characterization of CHI Chitinase
3.7. Hydrolytic Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tharanathan, R.N.; Kittur, F.S. Chitin-the undisputed biomolecule of great potential. Crit. Rev. Food Sci. Nutr. 2003, 43, 61–87. [Google Scholar] [CrossRef] [PubMed]
- Vyas, P.; Deshpande, M.V. Chitinase production by Myrotheciumver rucaria and its significance for fungal mycelia degradation. J. Gen. Appl. Microbiol. 1989, 5, 343–350. [Google Scholar] [CrossRef]
- Vyas, P.; Deshpande, M.V. Enzymatic hydrolysis of chitin by Myrothecium verrucaria Chitinase complex and its utilization to produce SCP. J. Gen. Appl. Microbiol. 1991, 3, 267–275. [Google Scholar] [CrossRef]
- Wang, S.L.; Lin, T.Y.; Yen, Y.H.; Liao, H.F.; Chen, Y.J. Bioconversion of shellfish chitin wastes for the production of Bacillus subtilis W-118 chitinase. Carbohydr. Res. 2006, 15, 2507–2515. [Google Scholar] [CrossRef] [PubMed]
- Songsiriritthigul, C.; Lapboonrueng, S.; Pechsrichuang, P.; Pesatcha, P.; Yamabhai, M. Expression and characterization of Bacillus licheniformis chitinase (ChiA), suitable for bioconversion of chitin waste. Bioresour. Technol. 2010, 11, 4096–4103. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, N.; Tewari, R.; Hoondal, G.S. Biotechnological aspects of chitinolytic enzymes: A review. Appl. Microbiol. Biotechnol. 2006, 6, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Hart, P.J.; Pfluger, H.D.; Monzingo, A.F.; Hollis, T.; Robertus, J.D. The refined crystal structure of an endochitinase from Hordeum vulgare L. seeds at 1.8 Å resolution. J. Mol. Biol. 1995, 2, 402–413. [Google Scholar] [CrossRef]
- Suzuki, K.; Taiyoji, M.; Sugawara, N.; Nikaidou, N.; Henrissat, B.; Watanabe, T. The third chitinase gene (chiC) of Serratia marcescens 2170 and the relationship of its product to other bacterial chitinases. Biochem. J. 1999, 343, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.J.; Chen, C.Y. High-level expression and characterization of two chitinases, ChiCH and ChiCW, of Bacillus cereus 28-9 in Escherichia coli. Biochem. Biophys. Res. Commun. 2005, 1, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Tsujibo, H.; Okamoto, T.; Hatano, N.; Miyamoto, K.; Watanabe, T.; Mitsutomi, M.; Inamori, Y. Family 19 chitinases from Streptomyces thermoviolaceus OPC-520: Molecular cloning and characterization. Biosci. Biotechnol. Biochem. 2000, 11, 2445–2453. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Sugawara, N.; Suzuki, M.; Uchiyama, T.; Katouno, F.; Nikaidou, N.; Watanabe, T. Chitinases A, B, and C1 of Serratia marcescens 2170 produced by recombinant Escherichia coli: Enzymatic properties and synergism on chitin degradation. Biosci. Biotechnol. Biochem. 2002, 5, 1075–1083. [Google Scholar] [CrossRef]
- Hao, Z.K.; Cai, Y.J.; Liao, X.R.; Liang, X.H.; Liu, J.Y.; Fang, Z.Y.; Hu, M.M.; Zhang, D.B. Chitinolyticbacter meiyuanensis SYBC-H1T, Gen. Nov., sp. Nov., a Chitin-degrading bacterium isolated from soil. Curr. Microbiol. 2011, 6, 1732–1738. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.K.; Cai, Y.J.; Liao, X.R.; Ge, X.Y.; Zhang, H.R.; Gao, F. Improvement of chitinase production from a newly isolated Chitinolyticbacter meiyuanensis SYBC-H1 using central composite design technique. Asian. J. Chem. 2011, 12, 5289–5294. [Google Scholar]
- Hao, Z.K.; Cai, Y.J.; Liao, X.R.; Zhang, X.L.; Fang, Z.Y.; Zhang, D.B. Optimization of nutrition factors on chitinase production from a newly isolated Chitiolyticbacter meiyuanensis SYBC-H1. Braz. J. Microbiol. 2012, 1, 177–186. [Google Scholar] [CrossRef]
- Chen, J.K.; Shen, C.R.; Yeh, C.H.; Fang, B.S.; Huang, T.L.; Liu, C.L. N-acetyl glucosamine obtained from chitin by chitin degrading factors in Chitinbacter tainanesis. Int. J. Mol. Sci. 2011, 2, 1187–1195. [Google Scholar] [CrossRef] [PubMed]
- Toharisman, A.; Suhartono, M.T.; Spindler-Barth, M.; Hwang, J.K.; Pyun, Y.R. Purification and characterization of a thermostable chitinase from Bacillus licheniformis Mb-2. World. J. Microb. Biot. 2005, 5, 733–738. [Google Scholar] [CrossRef]
- Brzezinska, M.S.; Jankiewicz, U.; Walczak, M. Biodegradation of chitinous substances and chitinase production by the soil actinomycete Streptomyces rimosus. Int. Biodeterior. Biodegrad. 2013, 84, 104–110. [Google Scholar] [CrossRef]
- Molinari, L.M.; Pedroso, R.B.; Oliveira-Scoaris, D.; Ueda-Nakamura, T.; Nakamura, C.V.; Dias Filho, B.P. Identification and partial characterisation of a chitinase from Nile tilapia, Oreochromis niloticus. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 2007, 1, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Pradeep, G.; Choi, Y.H.; Choi, Y.S.; Suh, S.E.; Seong, J.H.; Cho, S.S.; Bae, M.S.; Yoo, J.C. An extremely alkaline novel chitinase from Streptomyces sp. CS495. Process. Biochem. 2014, 2, 223–229. [Google Scholar] [CrossRef]
- Zhang, J.; Kopparapu, N.K.; Yan, Q.; Yang, S.; Jiang, Z. Purification and characterisation of a novel chitinase from persimmon (Diospyros kaki) with antifungal activity. Food. Chem. 2013, 2, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Flach, J.; Pilet, P.E.; Jolles, P. What’s new in chitinase research? Experientia 1992, 8, 701–716. [Google Scholar] [CrossRef]
- Gortari, M.C.; Hours, R.A. Biotechnological processes for chitin recovery out of crustacean waste: A mini-review. Electron. J. Biotechnol. 2013, 3. [Google Scholar] [CrossRef]
- Wortman, A.; Somerville, C.; Colwell, R. Chitinase determinants of Vibrio vulnificus: Gene cloning and applications of a chitinase probe. Appl. Environ. Microbiol. 1986, 1, 142–145. [Google Scholar]
- Nishizawa, Y.; Hibi, T. Rice chitinase gene: cDNA cloning and stress-induced expression. Plant Sci. 1991, 2, 211–218. [Google Scholar] [CrossRef]
- Tsujibo, H.; Orikoshi, H.; Tanno, H.; Fujimoto, K.; Miyamoto, K.; Imada, C.; Okami, Y.; Inamori, Y. Cloning, sequence, and expression of a chitinase gene from a marine bacterium, Altermonas sp. strain O-7. J. Bacteriol. 1993, 1, 176–181. [Google Scholar]
- García, I.; Lora, J.M.; dela-Cruz, J.; Benítez, T.; Llobell, A.; Pintor-Toro, J.A. Cloning and characterization of a chitinase (CHIT42) cDNA from the mycoparasitic fungus Trichoderma harzianum. Curr. Genet. 1994, 1, 83–89. [Google Scholar] [CrossRef]
- Boot, R.G.; Renkema, G.H.; Strijland, A.; van-Zonneveld, A.J.; Aerts, J.M. Cloning of a cDNA encoding chitotriosidase, a human chitinase produced by macrophages. J. Biol. Chem. 1995, 44, 26252–26256. [Google Scholar] [CrossRef]
- Yang, C.; Zhu, Y.; Magee, D.M.; Cox, R.A. Molecular cloning and characterization of the Coccidioides immitis complement fixation/chitinase antigen. Infect. Immun. 1996, 6, 1992–1997. [Google Scholar]
- Takaya, N.; Yamazaki, D.; Horiuchi, H.; Ohta, A.; Takagi, M. Cloning and characterization of a chitinase-encoding gene (chiA) from Aspergillus nidulans, disruption of which decreases germination frequency and hyphal growth. Biosci. Biotechnol. Biochem. 1998, 1, 60–65. [Google Scholar]
- Tsujibo, H.; Hatano, N.; Endo, H.; Miyamoto, K.; Inamori, Y. Purification and characterization of a thermostable chitinase from Streptomyces thermoviolaceus OPC-520 and cloning of the encoding gene. Biosci. Biotechnol. Biochem. 2000, 1, 96–102. [Google Scholar]
- Gao, B.; Allen, R.; Maier, T.; McDermott, J.P.; Davis, E.L.; Baum, T.J.; Hussey, R.S. Characterisation and developmental expression of a chitinase gene in Heterodera glycines. Int. J. Parasitol. 2002, 10, 1293–1300. [Google Scholar]
- Kurokawa, T.; Uji, S.; Suzuki, T. Molecular cloning of multiple chitinase genes in Japanese flounder, Paralichthys olivaceus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2004, 3, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Klemsdal, S.S.; Clarke, J.L.; Hoell, I.A.; Eijsink, V.G.; Brurberg, M.B. Molecular cloning, characterization, and expression studies of a novel chitinase gene (ech30) from the mycoparasite Trichoderma atroviride strain P1. FEMS Microbiol. Lett. 2006, 2, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Yang, Q. Cloning and expression of a novel chitinase chi58 from Chaetomium cupreum in Pichia pastoris. Biochem. Genet. 2009, 7–8, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.H.; Wang, Y.C.; Qi, X.T.; Yang, C.P. Cloning and characterization of a chitinase gene Lbchi31 from Limonium bicolor and identification of its biological activity. Mol. Biol. Rep. 2010, 5, 2447–2453. [Google Scholar] [CrossRef] [PubMed]
- Makkonen, J.; Jussila, J.; Kokko, H. The diversity of the pathogenic Oomycete (Aphanomyces astaci) chitinase genes within the genotypes indicate adaptation to its hosts. Fungal Genet. Biol. 2012, 8, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Liu, T.; Yang, Q. Cloning, expression and biocharacterization of of Cht5, the chitinase from the insect Ostrinia furnacalis. Insect Sci. 2013, 2, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Laribi-Habchi, H.; Bouanane-Darenfed, A.; Drouiche, N.; Pauss, A.; Mameri, N. Purification, characterization, and molecular cloning of an extracellular chitinase from Bacillus licheniformis stain LHH100 isolated from wastewater samples in Algeria. Int. J. Biol. Macromol. 2015, 72, 1117–1128. [Google Scholar] [CrossRef] [PubMed]
- Trudel, J.; Asselin, A. Detection of chitinase activity after polyacrylamide gel electrophoresis. Anal. Biochem. 1989, 2, 362–366. [Google Scholar] [CrossRef]
- Bradford, M. A rapid and sensitive method for the quantitation of micrograrn quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 2, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 5259, 680–685. [Google Scholar] [CrossRef]



| Purification Steps | Total Activity (U) | Total Protein (mg) | Specific Activity (U/mg) | Yield (%) | Purification Fold |
|---|---|---|---|---|---|
| Crude enzyme | 239.0 | 391.1 | 0.6 | 100 | 1 |
| Ammoniumsulfate | 161.1 | 106.0 | 1.5 | 67.4 | 2.5 |
| DEAE-cellulose | 90.3 | 24.5 | 3.9 | 37.8 | 6.0 |
| Sephadex G-100 | 50.0 | 8.0 | 6.2 | 20.9 | 10.2 |
| Chitinase | Microorganism | Optimum pH | Optima Temperature (°C) | Half-Life Time (°C) | Mw (kDa) | Ref. |
|---|---|---|---|---|---|---|
| CHI2 | C.meiyuanensis SYBC-H1 | 6.5 | 40 | 60 min (60 °C) | 42 | This study |
| Chi | Bacillus sp. NTCU2 | 7 | 60 | 30 min (60 °C) | 36.5 | [11] |
| Chi | Bacillus licheniformis MB-2 | 6 | 70 | 80 min (60 °C) | 67 | [8] |
| Chi | Streptomyces RC1071 | 8 | 40 | 60 min (60 °C) | 70 | [16] |
| Ch501 | Streptomyces sp. CS501 | 7 | 60 | 60 min (55 °C) | 43 | [10] |
| ChiA | Pseudoalteromonas sp. DL-6 | 8 | 20 | 60 min (40 °C) | 110 | [2] |
| Chemicals | 0 h | 24 h | 48 h | 72 h | 96 h | 120 h |
|---|---|---|---|---|---|---|
| No addition | 100 | 95.80 | 75.18 | 54.43 | 43.76 | 15.09 |
| Vc | 100 | 96.05 | 71.92 | 42.50 | 22.82 | 16.17 |
| VB6 | 100 | 91.18 | 64.73 | 58.45 | 41.47 | 15.43 |
| Coenzyme | 100 | 95.13 | 59.81 | 57.22 | 39.14 | 20.03 |
| Glutamine | 100 | 88.37 | 49.28 | 32.72 | 29.29 | 16.62 |
| Polyethylene | 100 | 76.62 | 60.45 | 56.36 | 41.36 | 36.91 |
| Cyclodextrin | 100 | 76.93 | 50.26 | 33.79 | 20.16 | 16.97 |
| Galactose | 100 | 96.59 | 36.06 | 16.30 | 13.12 | 12.34 |
| Soluble starch | 100 | 75.15 | 86.01 | 24.23 | 15.71 | 12.62 |
| EGTA | 100 | 94.87 | 84.60 | 62.91 | 63.06 | 44.50 |
| EDTA | 100 | 76.73 | 58.19 | 54.42 | 42.77 | 44.56 |
| Dithiothreitol | 100 | 94.46 | 66.25 | 52.08 | 32.13 | 18.22 |
| β-Mercaptoethanol | 100 | 127.50 | 101.33 | 86.54 | 73.83 | 46.47 |
| Gene Sources | Gene Name | Year | Reference |
|---|---|---|---|
| Vibrio vulnificus | pATW501, 502, 503 | 1986 | [23] |
| Rice | Rice Ch t | 1991 | [24] |
| Altermonas sp. strain O-7 | pCHI997 | 1993 | [25] |
| Trichoderma harzianum | CHIT42 | 1994 | [26] |
| Human | Chitotriosidase | 1995 | [27] |
| Coccidioides immitis CF | CF | 1996 | [28] |
| Aspergillus nidulans | ChiA | 1998 | [29] |
| Streptomyces thermoviolaceus OPC520 | Chi25 | 2000 | [30] |
| Heterodera glycines | designated Hg-chi-1 | 2002 | [31] |
| Paralichthys olivaceus | fChi1, fChi2 and fChi3 | 2004 | [32] |
| Trichoderma atroviride strain P1 | ech30 | 2006 | [33] |
| Pichia pastoris | chi58 | 2009 | [34] |
| Limonium bicolor | Lbchi31 | 2010 | [35] |
| Aphanomyces astaci | CHI1, CHI2 and CHI3 | 2012 | [36] |
| Ostrinia | OfCht5 | 2013 | [37] |
| Bacillus licheniformis LHH100 | ChiA-65 | 2015 | [38] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, Z.; Wu, H.; Yang, M.; Chen, J.; Xi, L.; Zhao, W.; Yu, J.; Liu, J.; Liao, X.; Huang, Q. Cloning, Expression and 3D Structure Prediction of Chitinase from Chitinolyticbacter meiyuanensis SYBC-H1. Int. J. Mol. Sci. 2016, 17, 825. https://doi.org/10.3390/ijms17060825
Hao Z, Wu H, Yang M, Chen J, Xi L, Zhao W, Yu J, Liu J, Liao X, Huang Q. Cloning, Expression and 3D Structure Prediction of Chitinase from Chitinolyticbacter meiyuanensis SYBC-H1. International Journal of Molecular Sciences. 2016; 17(6):825. https://doi.org/10.3390/ijms17060825
Chicago/Turabian StyleHao, Zhikui, Hangui Wu, Meiling Yang, Jianjun Chen, Limin Xi, Weijie Zhao, Jialin Yu, Jiayang Liu, Xiangru Liao, and Qingguo Huang. 2016. "Cloning, Expression and 3D Structure Prediction of Chitinase from Chitinolyticbacter meiyuanensis SYBC-H1" International Journal of Molecular Sciences 17, no. 6: 825. https://doi.org/10.3390/ijms17060825
APA StyleHao, Z., Wu, H., Yang, M., Chen, J., Xi, L., Zhao, W., Yu, J., Liu, J., Liao, X., & Huang, Q. (2016). Cloning, Expression and 3D Structure Prediction of Chitinase from Chitinolyticbacter meiyuanensis SYBC-H1. International Journal of Molecular Sciences, 17(6), 825. https://doi.org/10.3390/ijms17060825





