A Comparison of Fresh Frozen vs. Formalin-Fixed, Paraffin-Embedded Specimens of Canine Mammary Tumors via Branched-DNA Assay
Abstract
:1. Introduction
2. Results
2.1. RNA Quantification
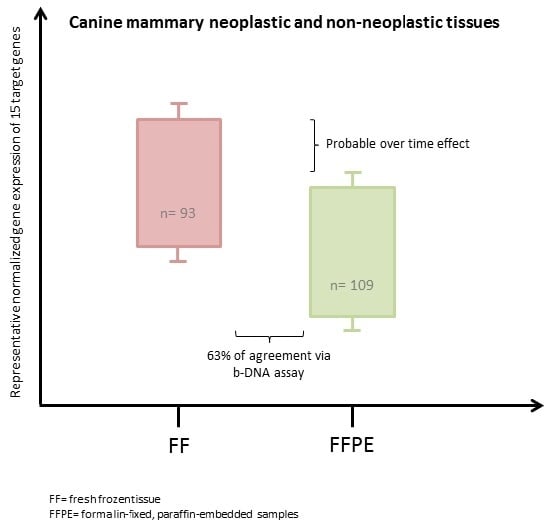
2.2. Gene Expression
2.2.1. Housekeeping Genes
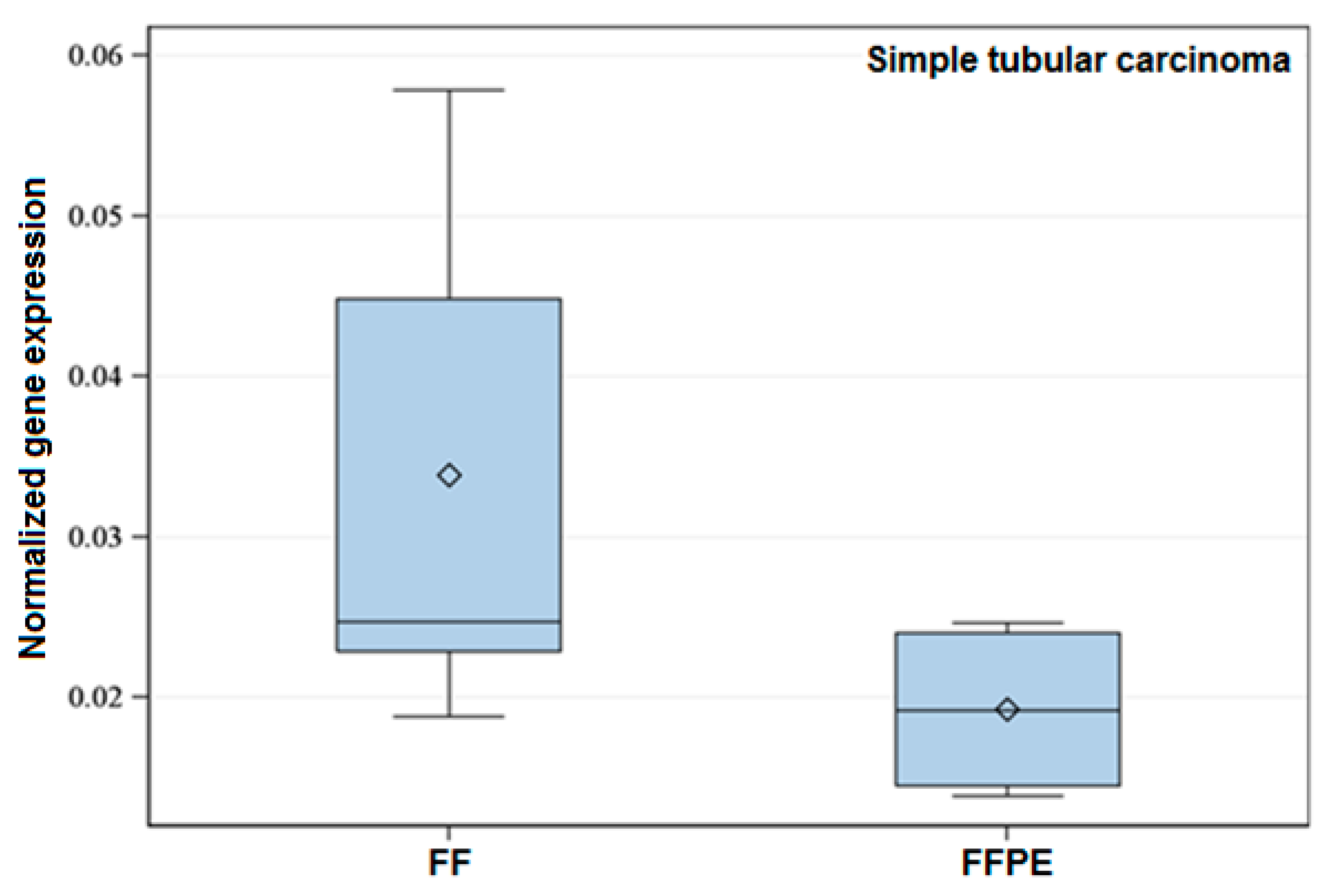
2.2.2. Target Genes and Histological Groups
2.3. Effect of Storage Time
2.4. Correlation between Fresh Frozen (FF) and Formalin-Fixed, Paraffin-Embedded (FFPE)
3. Discussion
4. Materials and Methods
4.1. Tissue Samples
4.1.1. Formalin-Fixed Paraffin Blocks
4.1.2. Fresh Frozen Tumor Samples
4.2. Nucleic Acid Isolation and Quantification
4.2.1. Formalin-Fixed Paraffin Samples
4.2.2. Fresh Frozen Samples
4.2.3. Target Genes
4.2.4. Housekeeping Genes
4.2.5. Luminex Technology (Branched-DNA (b-DNA) Assay)
4.2.6. Histological Classification
4.2.7. Data and Statistical Analysis
4.2.8. Effect of Storage Time
4.2.9. Correlation between FF and FFPE
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Merlo, D.F.; Rossi, L.; Pellegrino, C.; Ceppi, M.; Cardellino, U.; Capurro, C.; Ratto, A.; Sambucco, P.L.; Sestito, V.; Tanara, G.; et al. Cancer incidence in pet dogs: Findings of the Animal Tumor Registry of Genoa, Italy. J. Vet. Intern. Med. 2008, 22, 976–984. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; McCarron, P.; Parkin, D.M. The changing global patterns of female breast cancer incidence and mortality. Breast Cancer Res. 2004, 6, 229–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penland, S.K.; Keku, T.O.; Torrice, C.; He, X.; Krishnamurthy, J.; Hoadley, K.A.; Woosley, J.T.; Thomas, N.E.; Perou, C.M.; Sandler, R.S.; et al. RNA expression analysis of formalin-fixed paraffin-embedded tumors. Lab. Investig. 2007, 87, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Granato, A.; Giantin, M.; Ariani, P.; Carminato, A.; Baratto, C.; Zorzan, E.; Vascellari, M.; Bozzato, E.; Dacasto, M.; Mutinelli, F. DNA and RNA isolation from canine oncologic formalin-fixed, paraffin-embedded tissues for downstream “-omic” analyses: Possible or not? J. Vet. Diagn. Investig. 2014, 26, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Von Ahlfen, S.; Missel, A.; Bendrat, K.; Schlumpberger, M. Determinants of RNA quality from FFPE Samples. PLoS ONE 2007, 2. [Google Scholar] [CrossRef] [PubMed]
- Mittempergher, L.; de Ronde, J.J.; Nieuwland, M.; Kerkhoven, R.M.; Simon, I.; Emiel, J.; Wessels, L.F.A.; Van’t Veer, L.J. Gene expression profiles from formalin fixed paraffin embedded breast cancer tissue are largely comparable to fresh frozen matched tissue. PLoS ONE 2011, 6, e17163. [Google Scholar] [CrossRef] [PubMed]
- Perlmutter, M.A.; Best, C.J.; Gillespie, J.W.; Gathright, Y.; González, S.; Velasco, A.; Linehan, W.M.; Emmert-Buck, M.R.; Chuaqui, R.F. Comparison of snap freezing versus ethanol fixation for gene expression profiling of tissue specimens. J. Mol. Diagn. 2004, 6, 371–377. [Google Scholar] [CrossRef]
- Ramaswamy, S. Translating cancer genomics into clinical oncology. N. Engl. J. Med. 2004, 350, 1814–1816. [Google Scholar] [CrossRef] [PubMed]
- April, C.; Klotzle, B.; Royce, T.; Wickham-Garcia, E.; Boyaniwsky, T.; Izzo, J.; Cox, D.; Jones, W.; Rubio, R.; Holton, K.; et al. Whole-genome gene expression profiling of formalin-fixed, paraffin-embedded tissue samples. PLoS ONE 2009, 4, e8162. [Google Scholar] [CrossRef] [PubMed]
- Scicchitano, M.S.; Dalmas, D.A.; Bertiaux, M.A.; Anderson, S.M.; Turner, L.R.; Thomas, R.A.; Mirable, R.; Boyce, R.W. Preliminary comparison of quantity, quality, and microarray performance of RNA extracted from formalin-fixed, paraffin-embedded, and unfixed frozen tissue samples. J. Histochem. Cytochem. 2006, 54, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Werner, M.; Chott, A.; Fabiano, A.; Battifora, H. Effect of formalin tissue fixation and processing on immunohistochemistry. Am. J. Surg. Pathol. 2000, 24, 1016–1019. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, M.; Sedmak, D.; Jewell, S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am. J. Pathol. 2002, 161, 1961–1971. [Google Scholar] [CrossRef]
- Cassali, G.D.; Lavalle, G.E.; Ferreira, E.; Lima, A.E.; de Nardi, A.B.; Fernandes, C.G. Consensus for the diagnosis, prognosis and treatment of canine mammary tumors. Braz. J. Vet. Pathol. 2011, 4, 153–180. [Google Scholar]
- Bradley, W.H.; Eng, K.; Le, M.; Mackinnon, A.C.; Kendziorski, C.; Rader, J.S. Comparing gene expression data from formalin-fixed, paraffin embedded tissues and qPCR with that from snap-frozen tissue and microarrays for modeling outcomes of patients with ovarian carcinoma. BMC Clin. Pathol. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Tsongalis, G.J. Branched DNA technology in molecular diagnostics. Am. J. Clin. Pathol. 2006, 126, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, B.S.; Allen, A.N.; McLerran, D.F.; Vessella, R.L.; Karademos, J.; Davies, J.E.; Maqsodi, B.; McMaster, G.K.; Kristal, A.R. Evaluation of the branched-chain DNA assay for measurement of RNA in formalin-fixed tissues. J. Mol. Diagn. 2008, 10, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, H.; Yando, Y.; Nishio, M.; Zhang, Z.; Hamaguchi, M.; Mita, K.; Kobayashi, S.; Fujii, Y.; Iwase, H. Immunohistochemical evaluation of hormone receptor status for predicting response to endocrine therapy in metastatic breast cancer. Breast Cancer 2006, 13, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Gadzicki, D.; Evans, D.G.; Harris, H.; Julian-Reynier, C.; Nippert, I.; Schmidtke, J.; Tibben, A.; van Asperen, C.J.; Schlegelberger, B. Genetic testing for familial/hereditary breast cancer—Comparison of guidelines and recommendations from the UK, France, the Netherlands and Germany. J. Commun. Genet. 2011, 2, 53–69. [Google Scholar] [CrossRef] [PubMed]
- Queiroga, F.L.; Raposo, T.; Carvalho, M.I.; Prada, J.; Pires, I. Canine mammary tumours as a model to study human breast cancer: Most recent findings. In Vivo 2011, 25, 455–465. [Google Scholar] [PubMed]
- Goldschmidt, M.; Pena, L.; Rasotto, R.; Zappulli, V. Classification and grading of canine mammary tumors. Vet. Pathol. 2011, 48, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Cronin, M.; Pho, M.; Dutta, D.; Stephans, J.C.; Shak, S.; Kiefer, M.C.; Esteban, J.M.; Baker, J.B. Measurement of gene expression in archival paraffin-embedded tissues: Development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am. J. Pathol. 2004, 164, 35–42. [Google Scholar] [CrossRef]
- Nam, S.K.; Im, J.; Kwak, Y.; Han, N.; Nam, K.H.; Seo, A.N.; Lee, H.S. Effects of fixation and storage of human tissue samples on nucleic Acid preservation. Korean J. Pathol. 2014, 48, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Kim, T.; Kong, S.Y.; Kwon, S.; Bae, C.Y.; Choi, J.; Kim, C.H.; Lee, E.S.; Park, J.-K. Breast cancer diagnosis using a microfluidic multiplexed immunohistochemistry platform. PLoS ONE 2010, 5, e10441. [Google Scholar] [CrossRef] [PubMed]
- Matos, L.L.; Trufelli, D.C.; de Matos, M.G.; da Silva Pinhal, M.A. Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomark. Insights 2010, 5, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, C.; Karlsson, M.G. Effects of long-term storage on the detection of proteins, DNA, and mRNA in tissue microarray slides. J. Histochem. Cytochem. 2011, 59, 1113–1121. [Google Scholar] [CrossRef] [PubMed]
- Urdea, M.S.; Horn, T.; Fultz, T.J.; Anderson, M.; Running, J.A.; Hamren, S.; Ahle, D.; Chang, C.A. Branched DNA amplification multimers for the sensitive, direct detection of human hepatitis viruses. Nucleic. Acids Symp. Ser. 1991, 24, 197–200. [Google Scholar] [PubMed]
- Soutschek, J.; Akinc, A.; Bramlage, B.; Charisse, K.; Constien, R.; Donoghue, M.; Elbashir, S.; Geick, A.; Hadwiger, P.; Harborth, J.; et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 2004, 432, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Masuda, N.; Ohnishi, T.; Kawamoto, S.; Monden, M.; Okubo, K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999, 27, 4436–4443. [Google Scholar] [CrossRef] [PubMed]
- De Kok, J.B.; Roelofs, R.W.; Giesendorf, B.A.; Pennings, J.L.; Waas, E.T.; Feuth, T.; Swinkels, D.W.; Span, P.N. Normalization of gene expression measurements in tumor tissues: Comparison of 13 endogenous control genes. Lab. Investig. 2005, 85, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Simons, J.W. Direct comparison of GAPDH, β-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem. Biophys. Res. Commun. 1999, 259, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Kunth, K.; Hofler, H.; Atkinson, M.J. [Quantification of messenger RNA expression in tumors: Which standard should be used for best RNA normalization?]. Verh. Dtsch. Ges. Pathol. 1994, 78, 226–230. [Google Scholar] [PubMed]
- Majidzadeh, A.K.; Esmaeili, R.; Abdoli, N. TFRC and ACTB as the best reference genes to quantify Urokinase Plasminogen Activator in breast cancer. BMC Res. Notes 2011, 4. [Google Scholar] [CrossRef] [PubMed]
- Kilic, Y.; Celebiler, A.C.; Sakızlı, M. Selecting housekeeping genes as references for the normalization of quantitative PCR data in breast cancer. Clin. Transl. Oncol. 2014, 16, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Lyng, M.B.; Laenkholm, A.V.; Pallisgaard, N.; Ditzel, H.J. Identification of genes for normalization of real-time RT-PCR data in breast carcinomas. BMC Cancer 2008, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, L.; Fritsche, H.; Mennel, R.; Norton, L.; Ravdin, P.; Taube, S.; Somerfield, M.R.; Hayes, D.F.; Bast, R.C. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J. Clin. Oncol. 2007, 25, 5287–5312. [Google Scholar] [PubMed]
- Rivera, P.; Melin, M.; Biagi, T.; Fall, T.; Häggström, J.; Lindblad-Toh, K.; von Euler, H. Mammary tumor development in dogs is associated with BRCA1 and BRCA2. Cancer Res. 2009, 69, 8770–8774. [Google Scholar] [CrossRef] [PubMed]
- Mavaddat, N.; Antoniou, A.C.; Easton, D.F.; Garcia-Closas, M. Genetic susceptibility to breast cancer. Mol. Oncol. 2010, 4, 174–191. [Google Scholar] [CrossRef] [PubMed]

| Group | FF (n) | FFPE (n) |
|---|---|---|
| 1. Healthy canine mammarytissue | 13 | 4 |
| 2. Simple adenoma | 6 | 4 |
| 3. Intraductal papillary adenoma | 5 | 8 |
| 4. Complexa denoma | 17 | 11 |
| 5. Benign mixed tumor | 4 | 13 |
| 6. Simple tubular carcinoma | 12 | 13 |
| 7. Solid carcinoma | 8 | 7 |
| 8. Complex carcinoma | 13 | 11 |
| 9. Ductal carcinoma | 5 | 15 |
| 10. Carcinoma arising in a complex adenoma/mixed tumor | 10 | 8 |
| Target Gene | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Tp53 | – | – | * | * | – | * | * | * | * | * |
| Myc | * | – | * | * | – | * | * | – | – | – |
| HMGA1 | * | – | – | – | – | * | – | – | – | – |
| Pik3ca | – | – | – | – | – | – | – | – | – | – |
| Mcl1 | * | – | – | – | – | * | – | – | – | * |
| MAPK3 | – | – | – | * | – | * | – | – | * | * |
| FOXO3 | – | – | – | – | – | – | – | – | – | * |
| PTEN | – | – | * | – | * | * | – | * | * | – |
| GATA4 | – | x | – | – | * | * | – | – | – | x |
| PFDN5 | – | – | – | – | – | – | – | – | – | * |
| HMGB1 | * | * | * | * | * | * | * | * | * | * |
| MAPK1 | * | * | * | * | – | * | * | * | * | * |
| BRCA2 | – | x | – | * | – | – | * | – | – | – |
| BRCA1 | – | x | x | – | – | – | – | – | – | – |
| HMGA2 | * | – | x | * | * | * | x | – | – | – |
| Her2 | – | – | – | – | – | * | * | – | – | – |
| Gene | Accession Number |
|---|---|
| BRCA1 | NM_001013416 |
| BRCA2 | NM_001006653 |
| FOXO3 | NM_003639400 |
| PDFN5 | NM_001251949 |
| PTEN | NM_001003192 |
| Tp53 | NM_001003210 |
| GATA4 | NM_001048112 |
| Her2 | NM_001003217 |
| HMGA1 | NM_001003387 |
| HMGA2 | XM_005625590 |
| HMGB1 | NM_001002937 |
| MAPK1 | NM_001110800 |
| MAPK3 | NM_001252035 |
| Mcl1 | NM_001003016 |
| Myc | NM_001003246 |
| Pik3ca | XM_545208.4 |
| ACTB | XM_536888 |
| GAPDH | NM_001003142 |
| HPRT1 | NM_001003357 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lüder Ripoli, F.; Mohr, A.; Conradine Hammer, S.; Willenbrock, S.; Hewicker-Trautwein, M.; Hennecke, S.; Murua Escobar, H.; Nolte, I. A Comparison of Fresh Frozen vs. Formalin-Fixed, Paraffin-Embedded Specimens of Canine Mammary Tumors via Branched-DNA Assay. Int. J. Mol. Sci. 2016, 17, 724. https://doi.org/10.3390/ijms17050724
Lüder Ripoli F, Mohr A, Conradine Hammer S, Willenbrock S, Hewicker-Trautwein M, Hennecke S, Murua Escobar H, Nolte I. A Comparison of Fresh Frozen vs. Formalin-Fixed, Paraffin-Embedded Specimens of Canine Mammary Tumors via Branched-DNA Assay. International Journal of Molecular Sciences. 2016; 17(5):724. https://doi.org/10.3390/ijms17050724
Chicago/Turabian StyleLüder Ripoli, Florenza, Annika Mohr, Susanne Conradine Hammer, Saskia Willenbrock, Marion Hewicker-Trautwein, Silvia Hennecke, Hugo Murua Escobar, and Ingo Nolte. 2016. "A Comparison of Fresh Frozen vs. Formalin-Fixed, Paraffin-Embedded Specimens of Canine Mammary Tumors via Branched-DNA Assay" International Journal of Molecular Sciences 17, no. 5: 724. https://doi.org/10.3390/ijms17050724