Abstract
Drought is one of the important abiotic factors that adversely affects plant growth and production. The WRKY transcription factor plays a pivotal role in plant growth and development, as well as in the elevation of many abiotic stresses. Among three major groups of the WRKY family, the group IIe WRKY has been the least studied in floral crops. Here, we report functional aspects of group IIe WRKY member, i.e., CmWRKY10 in chrysanthemum involved in drought tolerance. The transactivation assay showed that CmWRKY10 had transcriptional activity in yeast cells and subcellular localization demonstrated that it was localized in nucleus. Our previous study showed that CmWRKY10 could be induced by drought in chrysanthemum. Moreover, the overexpression of CmWRKY10 in transgenic chrysanthemum plants improved tolerance to drought stress compared to wild-type (WT). High expression of DREB1A, DREB2A, CuZnSOD, NCED3A, and NCED3B transcripts in overexpressed plants provided strong evidence that drought tolerance mechanism was associated with abscisic acid (ABA) pathway. In addition, lower accumulation of reactive oxygen species (ROS) and higher enzymatic activity of peroxidase, superoxide dismutase and catalase in CmWRKY10 overexpressed lines than that of WT demonstrates its role in drought tolerance. Together, these findings reveal that CmWRKY10 works as a positive regulator in drought stress by regulating stress-related genes.
1. Introduction
Drought is the most prominent among the abiotic factors that decrease crop production worldwide. Reduced water availability is a limiting factor during the early stage of plant growth and development which reduces the photosynthesis rate; therefore, it has received significant attention from scientists [,]. Plants resist these stresses by stimulating stress-responsive genes and regulating physiological and molecular networks and signaling pathways against abiotic stresses [,,]. During unfavorable environmental conditions, abscisic acid (ABA) plays an important role by altering stress-responsive genes []. In plants, water-deficit conditions usually result in a high accumulation of ABA that leads to the production of H2O2 which serves as a signaling intermediate to promote stomatal closure [,]. Some of the well-known transcription factor families that play a critical role in abiotic stress especially under drought conditions in many plants include DREB, ERF, WRKY, MYC, MYB, bZIP, bHLH and NAC [,]. Each transcription factor family may trigger a set of genes or provoke a key gene to produce the requisite response needed by the plant.
As one of the largest families of transcription factors, the WRKY protein has a significant role in protecting against stresses such as drought, fluctuating temperatures and salinity. The typical features of WRKY transcription factors include the presence of one or two conserved WRKYGQK motifs at the N-terminal and the zinc finger motif at the C-terminal [,,]. The WRKY protein is involved in the regulation of evolutionary stages of plants such as dormancy, trichome development, seed formation, leaf death, embryogenesis, plant growth and metabolic pathways [,,,]. Different functions of WRKY have been identified and exclusive binding of this factor to the W-box of target gene promoters is well established and understood in many plant species []. Based on characteristic features of the WRKY motif, the entire family can be grouped into three clades (I, II, and III). On the basis of some unique functional motifs in addition to conserved ones, the group II has been categorized into subgroups referred to as IIa to IIe []. Many studies in Arabidopsis revealed the involvement of the WRKY family in drought-tolerance response. For example, AtWRKY63 (ABO3) gene used ABA-signaling pathway to repress the negative effects of drought []. Overexpression of AtWRKY57 improves drought tolerance by enhancing the ABA level in Arabidopsis []. Similarly, AtWRKY25 works in thermo-tolerance by manipulating the transcription of heat shock proteins while AtWRKY33 confers salt-stress tolerance [,]. In addition to this, low Pi level in Arabidopsis enhance the activity of AtWRKY6 to regulate PHOSPHATE1 expression []. AtWRKY18, AtWRKY40, and AtWRKY60 react to ABA and abiotic stress and AtWRKY70 and AtWRKY54 lessen osmotic stress by modifying the stomatal aperture [,]. Upregulation of OsWRKY30 works against drought tolerance by stimulating MAP kinases []. Overexpression of OsWRKY11 increases tolerance against heat and drought stress under the control of stress-inducible promoter HSP101. Similarly, upregulation of OsWRKY45 improves the resistance against salt and drought stresses [,,]. Overexpression of TaWRKY2 improves the tolerance against drought and salt stresses; however, TaWRKY19 not only alleviates the drought and salt stress but also improves the plant’s tolerance to low temperatures [,]. In addition, TaWRKY10 encourages tolerance to different abiotic stresses by modifying the osmotic balance []. Overexpression of the HvWRKY38 protects against drought and cold stress []. Upregulation of BdWRKY36 improves the tolerance against drought stress in transgenic tobacco plants []. The function of WRKY transcription factors has come to be understood due to many studies on the subject; nevertheless, it still requires more research into its role in the floral plant defense mechanism against stresses.
Chrysanthemum (Chrysanthemum morifolium) is an important cut flower and pot plant throughout the world. The main problems faced by the chrysanthemum grower are the different ranges of biotic and abiotic stresses [,]. Regarding abiotic stresses, one major issue that hampers the production and quality of the cut flowers of chrysanthemums is salinity and drought. Salinity, which is highly correlated with drought in a complex network, makes water unavailable to the chrysanthemum plants, resulting in a low uptake of water by plants from the soil. Subsequently, these conditions produce stunted plant growth, a low number of flower buds, a small flower size, a low number of fully bloomed flowers and flower sticks with a short shelf life [].
In a previous study, we identified many genes in the WRKY family that play a positive role in regulating the response to drought and salinity stresses []. The present study has focused on critically analyzing one gene, namely CmWRKY10 from the WRKY group IIe in response to drought conditions and to determine the signaling pathway associated with this gene.
2. Results
2.1. CmWRKY10 Belongs to WRKY Group IIe Family
In previous studies, we identified a set of different genes from the WRKY family in chrysanthemum using the RACE PCR technique. Among these genes, CmWRKY10 was found to be an ortholog of AtWRKY65, the open reading frame of which had a full length of 861 bp and encoded 287 amino acids. This amino acid sequence was used to search its ortholog sequence in the NCBI protein database using the BLASTp tool. The analysis yielded 18 ortholog sequences, which were further used for amino acid sequence alignment and phylogeny analysis as shown in Figure 1. Based on the sequences aligned with CmWRKY10, it was determined that CmWRKY10 belongs to the WRKY IIe group family and has characteristics of the transcription factor of AtWRKY65. High conservation with respect to the amino acid, the zinc-finger motif and the presence of the WRKY domain was deduced from the aligned sequence. CmWRKY10 was found to be very close to VvWRKY65, with a similarity of 56%. However, the identity with other orthologs, such as SlWRKY65, StWRKY65, NtWRKY65 and NsWRKY65 was lower than that with VvWRKY65, suggesting that CmWRKY10 has a wide variation and is quite different from the other members of the group.
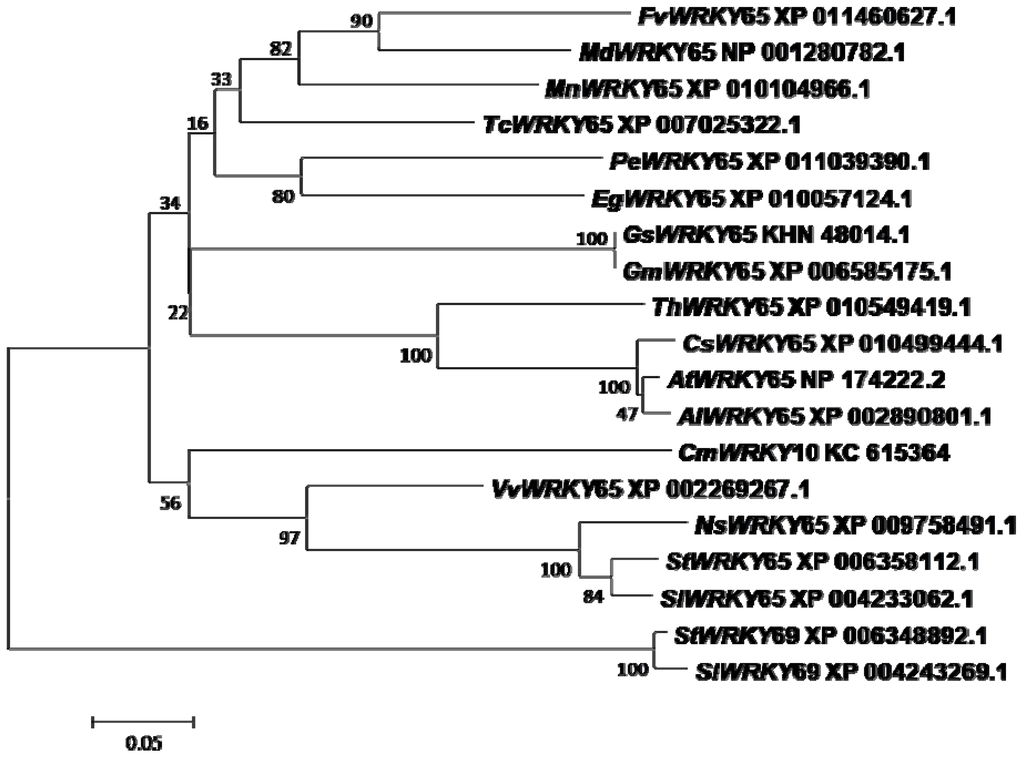
Figure 1.
A phylogenetic tree showing the relationship between CmWRKY10 and another plant’s WRKY proteins. At first, amino acid sequences were aligned with ClustalX software after that aligned file was used to construct the phylogenetic tree using neighbor-joining method with 1000 bootstraps in MEGA5 software.
2.2. CmWRKY10 Possess Transcriptional Activity in Yeast Cells
To determine whether CmWRKY10 possesses transcriptional activity, it was assayed by means of yeast one-hybrid system. As shown in Figure 2, the negative control pGBKT7 was not able to develop on SD/-His-Ade medium, unlike pGBKT7-CmWRKY10 and the positive control pCL1, which raised well on SD/-His-Ade medium and turned blue on SD+X-α-Gal medium, indicating that CmWRKY10 has transcriptional activity in yeast cell.

Figure 2.
An assessment of the transcription activation assay of CmWRKY10 using the yeast strain and a yeast one-hybrid system. The first column (a,c,e) shows the growth of transformants on SD/-His-Ade selection medium, where the growth of pCL1 (e) and empty vector pGBKT7 (c) represents the positive and negative control, respectively, representing the transcriptional activity of CmWRKY10; The second column (b,d,f) represents the growth of the same set of colonies in the presence of X-α-gal. Scale bars = 4 mm.
2.3. CmWRKY10 Localized in the Nucleus
Particle bombardment-mediated transient expression assay was used to discover the subcellular localization of CmWRKY10 in onion epidermal cells with the construct p35S::GFP-CmWRKY10 and a positive vector p35S::GFP. The GFP-CmWRKY10 fusion protein was localized in the nuclei of the onion epidermal cells (Figure 3). In contrast, the control GFP protein lacking CmWRKY10 was dispersed throughout the entire cell. These histological observations proved the subcellular localization of CmWRKY10 in the nucleus to function as a transcription factor.
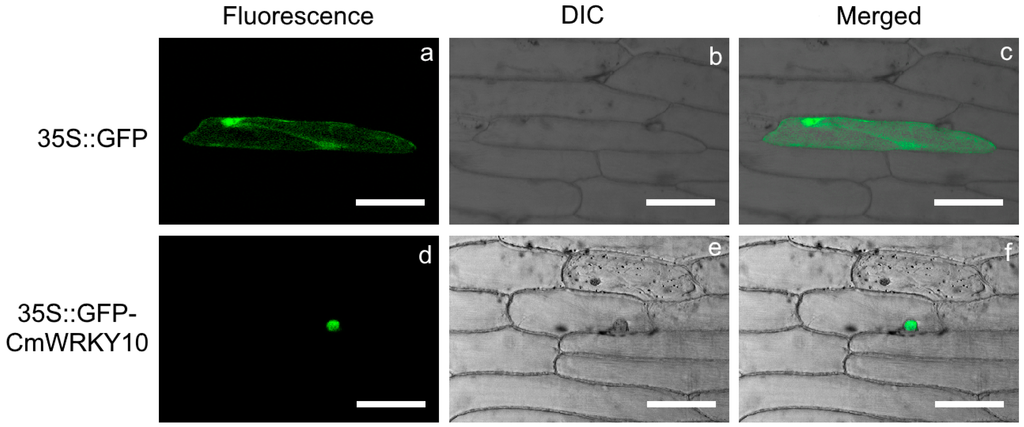
Figure 3.
Subcellular localization of CmWRKY10 protein in onion epidermal cell. GFP (upper panel) and GFP::CmWRKY10 (lower panel) fusion proteins were transiently expressed under the control of CaMV35S promoter and photographed with Zeizs Microsystem LSM730. Images were taken in dark field for green fluorescence (a,d), in differential interference contrast (DIC) (b,e) while the combination of green fluorescence and bright field (c,f) were also photographed. Scale bars = 50 μm.
2.4. CmWRKY10 Overexpression Increased Tolerance against Drought in Chrysanthemums
The chrysanthemum transgenic lines overexpressing CmWRKY10 were produced by Agrobacterium-mediated transformation. A total of 15 plants were able to regenerate on selection media which were further tested by PCR analysis for positive transformation. The relative expression levels of CmWRKY10 in all these transgenic lines were assessed using qRT-PCR. The expression level was notably higher in the overexpressed (OE) plants, ranging from 1.96- to 36-fold greater than those in the wild-type (WT) plants. The CmWRKY10 transcription levels in the lines OE-9, OE-12 and OE-14 were significantly higher than the rest of the OE lines, with expression levels of 20.74-, 32.32- and 36.71-fold greater than WT plants (Figure 4a). Hence, these three lines were selected for further drought-tolerance assessment. The plants from the selected lines were treated with 30% PEG6000 for 48 h and the results showed that the leaves of OE-9, OE-12 and OE-14 plants were less affected. Additionally, most plants survived the stress condition with less wilting of the leaves than the WT plants (Figure 4b). Following the recovery period, the survival rate of OE-9, OE-12 and OE-14 plants were 66.67%, 62.86% and 70.95%, respectively, as compared to WT plants which had the survival rate of 36.19% (Figure 4c). Overall, we can hypothesize that CmWRKY10 overexpression improves the drought tolerance in chrysanthemum.

Figure 4.
The relative expression level of CmWRKY10 gene in the overexpressed lines. Results are shown as means ± standard deviation (bars) of three biological repeats; (a) phenotypic comparison ofCmWRKY10 overexpressed lines(OE-9, OE-12 and OE-14) and wild-type ‘Jinba’ before and after the drought-stress treatment Scale bars = 60 mm (b). The survival rate of the wild-type plants and the overexpressed lines (OE-9, OE-12 and OE-14) after 5 days from a sample size of 65 plants of each individual genotype. Bars represent the standard deviation among three independent replicates (c).
2.5. CmWRKY10 Confers Drought Tolerance in Chrysanthemums through Abscisic Acid (ABA) Pathway
To identify the regulatory process of CmWRKY10 against drought stress, the expression of the ABA-responsive genes DREB1A, DREB2A, NCED3A, NCED3B and CuZnSOD were assessed. The results showed that DREB1A and DREB2A were upregulated in the OE lines after 1 h of stress treatment and remained at a high level at all of the time points. In contrast, low expression of NCED3A was found in OE lines at 1 h of stress application which sharply increased to reach the maximum expression at 12 h and remained significantly higher than WT plants for the rest of time intervals. In addition, the expressions of NCED3B and CuZnSOD in WT plants were significantly lower than that in the OE lines following all the treatments (Figure 5). All these results support our hypothesis that the CmWRKY10 uses ABA-signaling pathway to confer tolerance against drought in chrysanthemum.
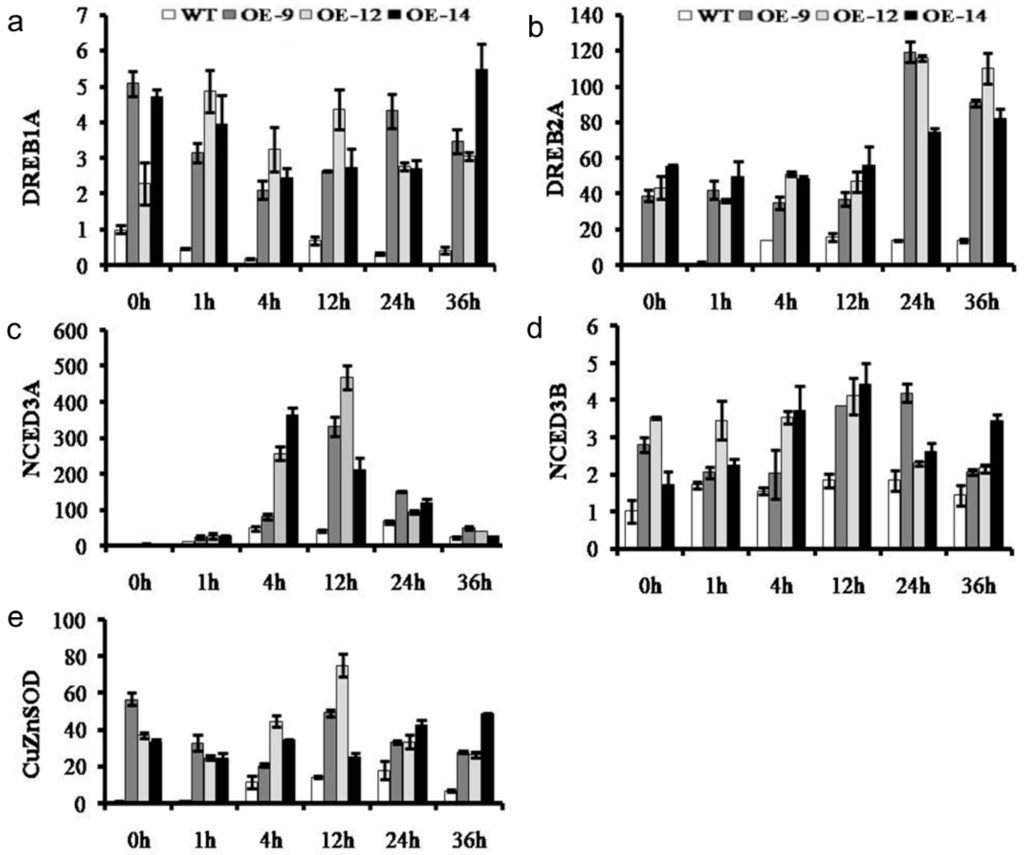
Figure 5.
The relative expression of abscisic acid (ABA)-responsive genes DREB1A (a), DREB2A (b), NCED3A (c), NCED3B (d) and CuZnSOD (e) in wild-type and transgenic lines at various time points (0, 1, 4, 12, 24 and 36 h). Bars represent the standard deviation among three independent replicates.
2.6. Overexpression of CmWRKY10 Reduces Recactive Oxygen Species (ROS) Accumulation due to the Enhanced Activity of Superoxide Dismutase (SOD), Peroxide Dismutase (POD) and Catalase (CAT) under Drought Stress
To obtain better insight into the function of CmWRKY10 in chrysanthemum plants under water-deficit conditions, we observed the accumulation of SOD, POD, CAT and ROS in the leaves of CmWRKY10 OE lines and WT plants at various time points i.e., 0, 1, 4, 12, 24and 36 h. Results shown in Figure 6 reveal that overexpression of CmWRKY10 lessened the ROS accumulation in OE lines at all of the time points, whereas it increased in WT plants after 1h of stress treatment and remained significantly higher at nearly all of the time points. The results also showed that the activity of three main antioxidant enzymes i.e., SOD, POD and CAT, had increased in OE lines as compared to WT. The value of SOD activity in the WT plants remained at 0.75 U for all of the time points whereas in the OE lines a high accumulation of SOD was observed at all of the time points. In the case of POD, no significant change in the enzyme activity was observed in WT plant during the stress treatment period; however, in all the OE lines, the concentration of POD was significantly higher than WT at all the time points. In OE lines, POD activity continuously increased after treatment and reached its maximum values at 36h post treatment. Results regarding CAT accumulation revealed that in WT plants there were no significant changes observed for CAT concentration during drought treatment as compared to OE lines in which a high concentration of CAT was measured against all time points after drought treatment. Taken together, these results provide strong evidence that CmWRKY10 decreased ROS accumulation by means of enhanced activity of SOD, POD and CAT to cope with drought conditions.
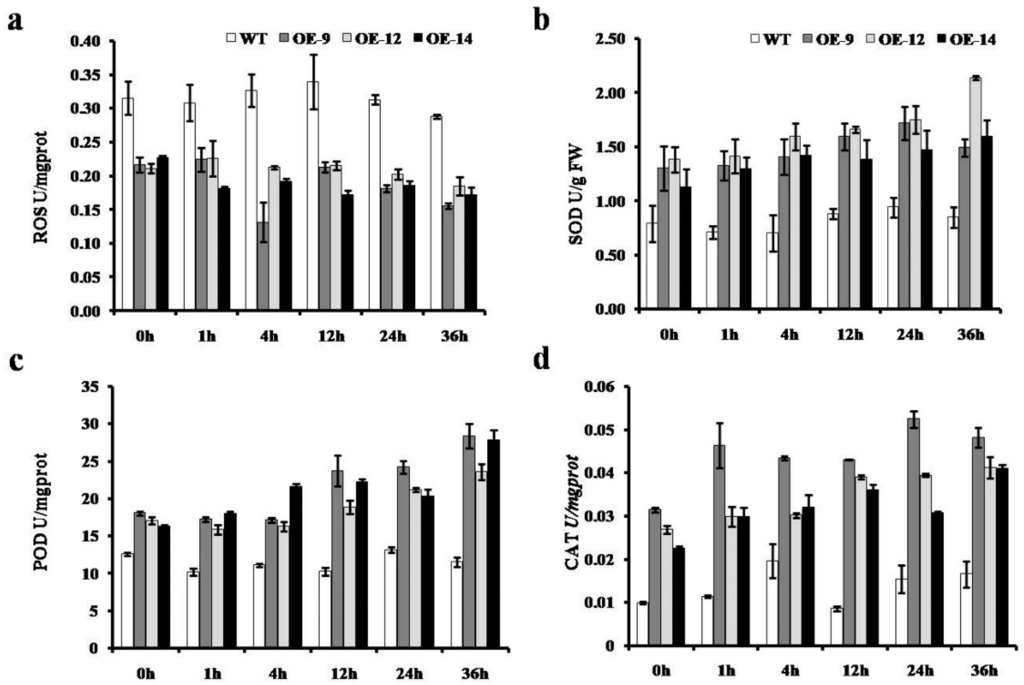
Figure 6.
Activities of reactive oxygen species (ROS) (a), SOD (b), POD (c) and CAT (d) in the wild-type and overexpressed lines (OE-9, OE-12 and OE-14) under normal and drought conditions at various time points (0, 1, 4, 12, 24 and 36 h). Bars represent the standard deviation among three independent replicates.
3. Discussion
The chrysanthemum is one of the most important cut flower ornamental plants. Like all other ornamental and crop plants, its growth and productivity are prone to many abiotic and biotic stresses. Among the abiotic factors that critically hamper its growth and deteriorate its quality are salinity and drought. Both of these stresses interact with each other in a complex manner to produce very negative effects on plant growth and the reproduction phase. Therefore, achieving tolerance to these stresses remains a top priority for breeders []. Accordingly, this study reported the involvement of one WRKY family gene CmWRKY10 in drought tolerance in chrysanthemum. Our results showed that CmWRKY10 relates to group II of the WRKY family and is a homolog to WRKY65 in Arabidopsis. The phylogenetic examination of the WRKY proteins revealed that the WRKY family is divided into three groups (I, II and III) with group II being further separated into five subgroups (a to e) [,]. Differential expression of the WRKY transcription factor critically governs plant developmental and other physiological activities []. A large number of WRKY genes and their functions have been well known since the identification of the first WRKY gene, “SPF1”, in sweet potatoes (Ipomoea batatas) [].
The subcellular localization of GFP-CmWRKY10 depicted that CmWRKY10-tagged GFP signals were concentrated only in the nucleus which proved the basis of its functionality as it has been demonstrated in many studies that the transcription factor must reside in the nucleus to perform its activity [,,]. Transcriptional activation analysis illustrated that CmWRKY10 has transcriptional activity in yeast cells, as reported by Sun et al. []. The high expression of CmWRKY10 authenticates the involvement of this transcription factor in alleviating drought stress in chrysanthemum. The analysis of CmWRKY10 overexpressed lines showed that there is a positive correlation between the expression of CmWRKY10 and survival rate of the plants when subjected to drought stress (Figure 4b). These findings further corroborate our hypothesis and is in accordance with previous studies. For example, overexpression of wheat TaWKRY2 and TaWRKY19 in Arabidopsis improved drought tolerance through regulation of the downstream genes [].
In the literature, there is convincing evidence of ABA’s role in abiotic stresses especially for drought stress. It accumulates in plants under osmotic stress and drought conditions []. Different transcription profiles which are involved in regulating abiotic stresses such as less water availability and low temperature in plants are controlled by ABA []. Our results revealed that CmWRKY10 confers drought tolerance in chrysanthemum through the ABA-signaling pathway. The higher expression of the ABA-related gene in the OE plants as compared to WT plants validated this hypothesis. These results are in accordance with previous findings that WRKY63/ABO3 regulates ABA contents in Arabidopsis for drought tolerance in the same way as rice, whereby OsWRKY45 interacts with ABA-signaling pathway genes to cope with the stress []. In addition, this same mechanism was observed for soybean WRKY20 when expressed in Arabidopsis. []. Moreover, it is well understood that DREB genes, especially DREB2, control the drought and salinity-signaling pathway []. These genes subsequently regulate key physiological functions involved in the tolerance mechanism []. NCED is another important enzyme in the synthesis of ABA and it also regulates the drought-stress response []. In contrast with the above results, our study showed that the overexpression of CmWRKY10 improves the survival of chrysanthemum plants more than WT plants by upregulating the ABA-biosynthesis genes DREB1A, DREB2A, NCED3A, NCED3B and CuZnSOD.
Moreover, the accumulation of ABA enhanced ROS activity causing many physiological and metabolic changes in plants to allow them to cope with the stress [,]. It has previously been proven in transgenic tobacco plants that wheat TaWRKY10 improved the level of proline and soluble sugar contents in cells by reducing the ROS accumulation to confer tolerance against drought stress []. Plants have developed intricate antioxidants such as SOD, POD and CAT to keep them safe from oxidative loss, produced by high accumulation of ROS []. In plants, ROS homeostasis is furnished by a complex system of antioxidants which not only scavenge ROS but also protect cells from oxidative damage [,]. Some recent studies have shed light on the activity of these antioxidants in relation to WRKY transcription factors for abiotic stresses, especially for drought and salinity. For instance, overexpressing of DgWRKY3 gene in tobacco resulted in enhanced activity of SOD, POD and CAT to minimize the salt-stress effect []. A similar function is executed by the SlWRKY gene in tobacco which is effective in both the stresses i.e., drought and salt []. In addition, overexpression of the BdWRKY36 gene in tobacco confers increased tolerance against drought stress due to the increased activity of SOD, POD and CAT []. Furthermore, we studied the ROS accumulation in these plants and the outcomes demonstrated that the tolerance of overexpressed lines against drought stress was enhanced because of reduced ROS contents compared with that in WT plants (Figure 6). It is important for the plant to keep the low ROS level to protect itself from oxidative stress and achieves this mostly by triggering the antioxidant activities []. Keeping this in view in the present study, the levels of some key antioxidant enzymes like SOD, POD and CAT were measured at different time intervals and the results indicated that the CmWRKY10 OE plants had enhanced SOD, POD and CAT activity compared to WT plants when subjected to water-stress conditions (Figure 6). These results helped us to speculate that it is the CmWRKY10 that scavenges the ROS so that OE plants may work more efficiently than WT plants. Collectively, all these results demonstrated that overexpression of the CmWRKY10 gene may activate the antioxidant defense system resulting in less ROS-mediated injury of transgenic plants under drought stress (Figure 7).
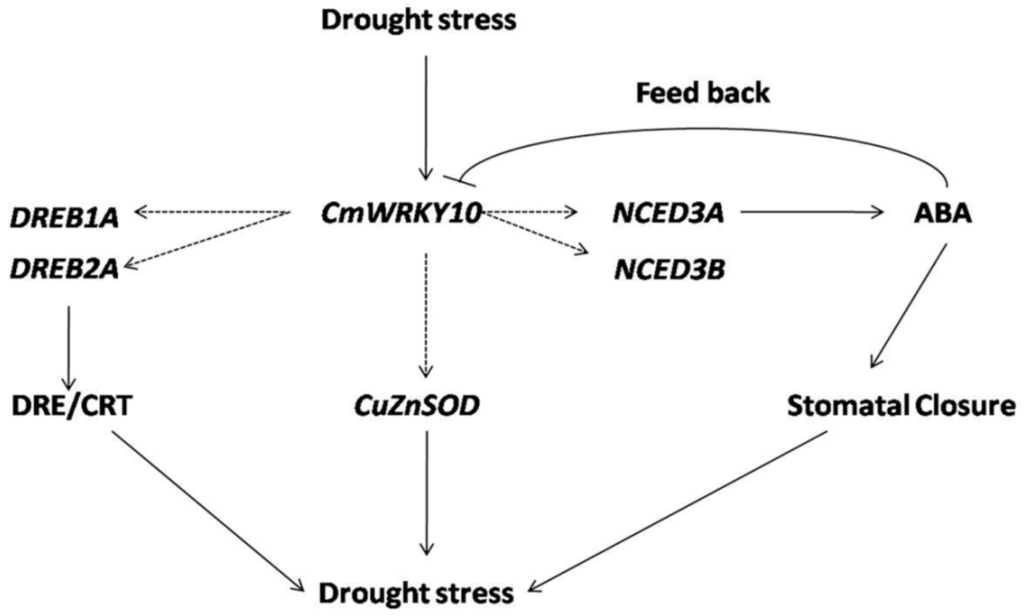
Figure 7.
A model of the interaction between CmWRKY10 and ABA-responsive genes under drought conditions. Solid arrows define the proved interactions while dashed arrows present the proposed interactions based on the results of present study.
In conclusion, our results show that the CmWRKY10 from chrysanthemum is a member of group IIe of the WRKY family, which is a positive regulator of drought tolerance. Functional analysis of CmWRKY10 proved that it causes transcriptional activity by regulating the transcription level of stress-response genes, especially ABA-signaling pathway genes. Future research will be focused on the potential target genes of CmWRKY10 to elucidate the molecular mechanisms associated with CmWRKY10 mediated stress tolerance.
4. Materials and Methods
4.1. Plant Material and Growth Conditions
For this experiment, we used the “Jinba” cultivar of chrysanthemum from the Chrysanthemum Germplasm Conservation Center, Nanjing Agricultural University, China. A soil and vermiculite mixture (1:1 v/v) was used for propagation of identical cuttings of the plant in a greenhouse under standard growth conditions of light and temperature (day/night; 14/10 h, light intensity 50 µmol·m−2·s−1 and 25/18 °C) with 70% relative humidity.
4.2. Sequence Analysis of CmWRKY10
The amino acid sequence from the previous study [] was used to search for its homolog protein sequences in different dicot species in the NCBI protein database using the BLASTp online tool. All of the sequences with more than 90% coverage were obtained and the sequence alignment of CmWRKY10 was performed along with that of different WRKY orthologs of different species using ClustalX software. For the construction of phylogenetic trees neighbor-joining method with 1000 bootstrap was applied, using MEGA5 software.
4.3. Transcriptional Activation Analysis in Yeast Cell
The CmWRKY10 coding sequence without the stop codon was amplified using a Phusion® High Fidelity PCR Kit (New England Biolabs, Ipswich, MA, USA) using the primer pair CmWRKY10-GATE-SAL-F/CmWRKY10-GATE-NOT-R (Table 1). The amplified product was inserted in the pENTR™1A vector (Invitrogen, Carlsbad, CA, USA) between Sal I and Not I cloning sites for ligation and recombinant vector was confirmed by sequencing []. Both pENTR™1A-CmWRKY10 and pDEST-GBKT7 were recombined to pDEST-GBKT7-CmWRKY10 using the LR Clonase™ II enzyme mix (Invitrogen, Carlsbad, CA, USA). The construct pDEST-GBKT7-CmWRKY10, pCL1 as positive control, and empty vector pDEST-GBKT7 as negative control were transferred to Saccharomyces cerevisiae yeast strain Y2HGold (Clontech, Mountain View, CA, USA) according to the producer’s directions. Collection of the transformants was conducted by either pDEST-GBKT7-CmWRKY10 or pDEST-GBKT7 using SD/-Trp medium; however, pCL1 was grown in SD/-Leu medium. Yeast cells transformed with pCL1 as the positive control were grown on SD/-His-Ade medium. However, yeast cells with pDEST-GBKT7 could not form a colony on this medium. The clones were then placed on SD/-His-Ade medium supplemented with X-α-gal and incubated at 30 °C for three days before monitoring their ability to spread [].

Table 1.
The primer names and sequences used in this study.
4.4. Subcellular Localization
Onion epidermal cells were used to detect the subcellular localization of CmWRKY10. To construct the GFP::CmWRKY10, the complete ORF of CmWRKY10 was cloned behind the GFP sequence in a pMDC43 vector using the LR Clonase™ II enzyme mixture (Invitrogen). The resulting p35S::GFP-CmWRKY10 construct and empty pMDC43 vector were delivered into onion epidermal cell layers by applying the gold particle bombardment technique []. The bombarded onion epidermal layers were incubated under dark conditions for at least 20 h at 24 °C prior to microscopic observation. The GFP signals from onion epidermal layers were detected and visualized by confocal laser microscope.
4.5. Chrysanthemum Transformation and Generation of Transgenic Lines
Initially, the complete ORF of CmWRKY10 was inserted in the plant expression vector pMDC32 to construct the p35S::CmWRKY10 vector using the LR Clonase™ II enzyme mix (Invitrogen). Then, this p35S::CmWRKY10 vector was transmuted into EHA105 strain of Agrobacterium tumefaciens by applying freeze-thaw transformation technique and following the protocol documented by Li et al. []. Briefly, young leaves of the chrysanthemum “Jinba” were cut into 0.5 cm diameter pieces and cultured on Murashige and skoog media that contained 8 mg·g−1 hygromycine for the selection of positive transformants []. The sprouting calli were differentiated into small plantlets following the protocol reported by Cui et al. []. The successfully regenerated plantlets were then shifted to a greenhouse for further growth under optimal environmental conditions.
The RNA was extracted from OE lines and WT plants after regeneration using RNAiso reagent (TaKaRa, Tokyo, Japan) and RNase-free DNase I (TaKaRa) was used to remove all the traces of DNA. The purified RNA was reverse-transcribed utilizing M-MLV (TaKaRa) enzyme using PCR. For the quantification of CmWRKY10 expression level, quantitative real-time PCR (qRT-PCR) assay was performed utilizing SYBR® Green reaction kit (TaKaRa) and CmWRKY10 specific primer pair (CmWRKY10-DL-F/R; Table 1). For the normalization of qRT-PCR data chrysanthemum reference gene CmEF1α was used. The outcomes of the transcription (having three biological replicates) were calculated using the 2–ΔΔCt method [].
4.6. Drought Stress Tolerance Assay for Transgenic Lines
A set of 65 cuttings at the 8–10 leaf stage of both transgenic and WT plants was propagated in a 30% PEG6000 solution for 48 h and the roots were then washed and kept in clean water. The plant recovery data were calculated after 5 days []. The samples (three biological replications) were collected at specific time points, i.e., 0, 1, 4, 12, 24 and 36 h, after the drought treatment. The samples were kept at −80 °C until further use and RNA isolation.
4.7. Expression Profiling of Drought Stress-Related Genes in Overexpressed (OE) Lines of CmWRKY10
To determine the regulatory mechanism of CmWRKY10 against drought stress, the expression levels of the different stress-responsive genes (DREB1A, DREB2A, NCED3A, NCED3B and CuZnSOD) were assessed by qRT-PCR. The sequences of all the primer pairs used in this study are listed in Table 1.
4.8. Measurements of Physiological–Biochemical Parameters
The ROS content and the activities of POD, CAT and SOD were measured by spectrophotometer utilizing commercial kit (E004, A084-3, A007-1 and A001-4, Jiancheng, Nanjing, China) following the manufacturer’s instructions. Briefly, approximately 0.5 g of chrysanthemum leaves were homogenized in 2 mL of extraction buffer supplemented with 0.1 M phosphate buffer (pH 7.8). Supernatant was separated from samples by centrifuge at 10,000× g at 4 °C for 15 min for determination of SOD, POD and CAT enzyme activities.
4.9. Statistical Analysis
All of the data recorded for the different physiological parameters were analyzed using one-way analysis of variance (ANOVA) to categorize the difference between the treatments with the least significant difference (LSD) test. For all the statistical analyses the software SPSS v17.0 (SPSS Inc., Chicago, IL, USA) was used.
Acknowledgments
This study was funded by the 863 project by Ministry of Science and Technology of China (2011AA100208), National Natural Science Foundation of China (31471913, 31501792), the Natural Science Fund of Jiangsu Province (BK20150657), 333 High-Level Personnel Training Project of Jiangsu Province (BRA2015315), the China Postdoctoral Science Foundation (2014M561673, 2015T80564), the Fundamental Research Funds for the Central Universities (KYTZ201401) and the project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author contributions
Aiping Song conceived and designed the experiments. Muhammad Abuzar Jaffar, Chen Liu and Qingqing Fan performed the experiments. Muhammad Faheem analyzed the data. Sumei Chen and Jiafu Jiang contributed reagents/materials/analysis tools. Fadi Chen drafted the manuscript. All authors read and approved the final manuscript.
Conflict of interest
The authors declare no conflict of interest.
References
- Shao, H.B.; Chu, L.Y.; Jaleel, C.A.; Manivannan, P.; Panneerselvam, R.; Shao, M.A. Understanding water deficit stress-induced changes in the basic metabolism of higher plants-biotechnologically and sustainably improving agriculture and the ecoenvironment in arid regions of the globe. Crit. Rev. Biotechnol. 2009, 29, 131–151. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Rabara, R.C.; Rushton, P.J. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 2014, 239, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Hu, W.; Zhou, R.; Wang, L.; Wang, X.; Wang, Q.; Feng, Z.; Li, Y.; Qiu, D.; He, G. The Brachypodium distachyon BdWRKY36 gene confers tolerance to drought stress in transgenic tobacco plants. Plant Cell Rep. 2015, 34, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Cai, H.; Zhai, H.; Luo, X.; Wang, Z.; Cui, L.; Bai, X. Overexpression of Glycinesoja WRKY20 enhances both drought and salt tolerance in transgenic alfalfa (Medicago sativa L.). Plant Cell Tissue Organ Cult. (PCTOC) 2014, 118, 77–86. [Google Scholar] [CrossRef]
- Jiang, S.C.; Mei, C.; Liang, S.; Yu, Y.T.; Lu, K.; Wu, Z.; Wang, X.F.; Zhang, D.P. Crucial roles of the pentatricopeptide repeat protein SOAR1 in Arabidopsis response to drought, salt and cold stresses. Plant Mol. Biol. 2015, 88, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Jakab, G.; Ton, J.; Flors, V.; Zimmerli, L.; Métraux, J.P.; Mauch-Mani, B. Enhancing Arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol. 2005, 139, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.K.; Gong, Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010, 63, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K.; Agarwal, P.; Reddy, M.; Sopory, S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006, 25, 1263–1274. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 2012, 10, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Zhang, L.B.; Guo, D.; Li, C.Z.; Peng, S.Q. Identification and expression profiles of the WRKY transcription factor family in Ricinus communis. Gene 2012, 503, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.F.; Chen, J.; Chen, Y.F.; Wu, L.J.; Xie, D.X. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012, 19, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, Y.; Jiang, J.; Chen, S.; Chen, F.; Guan, Z.; Fang, W. The constitutive expression of Chrysanthemum dichrum ICE1 in Chrysanthemum grandiflorum improves the level of low temperature, salinity and drought tolerance. Plant Cell Rep. 2012, 31, 1747–1758. [Google Scholar] [CrossRef] [PubMed]
- Li, J.B.; Luan, Y.S.; Jin, H. The tomato SlWRKY gene plays an important role in the regulation of defense responses in tobacco. Biochem. Biophys. Res. Commun. 2012, 427, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009, 69, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Fu, Q.; Huang, W.; Yu, D. Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep. 2009, 28, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.L.; Chen, F.D.; Chen, S.M. Establishment of regeneration and transformation system of ground-cover chrysanthemum Yuhuaxunzhang. J. Nanjing Agric. Univ. 2009, 32, 40–46. [Google Scholar]
- Chen, H.; Lai, Z.; Shi, J.; Xiao, Y.; Chen, Z.; Xu, X. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 2010, 10, 281. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Besseau, S.; Törönen, P.; Sipari, N.; Kollist, H.; Holm, L.; Palva, E.T. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 2013, 200, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Liu, C.; Zhang, Y.; Meng, X.; Zhou, X.; Chu, C.; Wang, X. OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol. Biol. 2012, 80, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Yu, D. Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 2009, 65, 35–47. [Google Scholar] [CrossRef]
- Wu, X.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009, 28, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Song, A.; Gao, C.; Jiang, J.; Chen, S.; Fang, W.; Zhang, F.; Chen, F. The over-expression of a chrysanthemum WRKY transcription factor enhances aphid resistance. Plant Physiol. Biochem. 2015, 95, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Deng, P.; Chen, L.; Wang, X.; Ma, H.; Hu, W.; Yao, N.; Feng, Y.; Chai, R.; Yang, G. A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS ONE 2013, 8, e65120. [Google Scholar] [CrossRef] [PubMed]
- Mare, C.; Mazzucotelli, E.; Crosatti, C.; Francia, E.; Cattivelli, L. HvWRKY38: A new transcription factor involved in cold-and drought-response in barley. Plant Mol. Biol. 2004, 55, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, J.A.T. Chrysanthemum: Advances in tissue culture, cryopreservation, postharvest technology, genetics and transgenic biotechnology. Biotechnol. Adv. 2003, 21, 715–766. [Google Scholar] [CrossRef]
- Song, A.; An, J.; Guan, Z.; Jiang, J.; Chen, F.; Lou, W.; Fang, W.; Liu, Z.; Chen, S. The constitutive expression of a two transgene construct enhances the abiotic stress tolerance of chrysanthemum. Plant Physiol. Biochem. 2014, 80, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Song, A.; Gao, C.; Wang, L.; Wang, Y.; Sun, J.; Jiang, J.; Chen, F.; Chen, S. Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Rep. 2015, 34, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Li, P.; Jiang, J.; Chen, S.; Li, H.; Zeng, J.; Shao, Y.; Zhu, L.; Zhang, Z.; Chen, F. Phylogenetic and transcription analysis of chrysanthemum WRKY transcription factors. Int. J. Mol. Sci. 2014, 15, 14442–14455. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. BBA-Gene Regul. Mech. 2012, 1819, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant. Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.L.; Zhong, M.; Li, S.; Pan, Y.Z.; Jiang, B.-B.; Jia, Y.; Zhang, H.Q. Overexpression of a chrysanthemum transcription factor gene DgWRKY3 in tobacco enhances tolerance to salt stress. Plant Physiol. Biochem. 2013, 69, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.F.; Wei, W.; Zhou, Q.Y.; Tian, A.G.; Hao, Y.J.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, Z.B.; Zhang, J.S. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012, 35, 1156–1170. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Bai, X.; Sun, X.; Zhu, D.; Liu, B.; Ji, W.; Cai, H.; Cao, L.; Wu, J.; Hu, M. Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J. Exp. Bot. 2013, 64, 2155–2169. [Google Scholar] [CrossRef] [PubMed]
- Sazegari, S.; Niazi, A.; Ahmadi, F.S. A study on the regulatory network with promoter analysis for Arabidopsis DREB-genes. Bioinformation 2015, 11, 101. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Kamei, A.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Molecular responses to drought, salinity and frost: Common and different paths for plant protection. Curr. Opin. Biotechnol. 2003, 14, 194–199. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, J. Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J. Exp. Bot. 2002, 53, 2401–2410. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Jiang, G.; Ye, N.; Chu, Z.; Xu, X.; Zhang, J.; Zhu, G. A key ABA catabolic gene, OsABA8ox3, is involved in drought stress resistance in rice. PLoS ONE 2015, 10, e0116646. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.-S.; Liu, J.H.; Chen, X.J. Overexpression of PtrABF gene, a bZIP transcription factor isolated from Poncirus trifoliata, enhances dehydration and drought tolerance in tobacco via scavenging ROS and modulating expression of stress-responsive genes. BMC Plant Biol. 2010, 10, 230. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Suzuki, N.; Ciftci, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Shigeoka, S. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 2011, 155, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Song, A.; Xin, J.; Chen, S.; Jiang, J.; Wang, Y.; Li, X.; Chen, F. CmWRKY15 facilitates Alternaria tenuissima infection of chrysanthemum. PLoS ONE 2015, 10, e0143349. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Song, A.; Li, P.; Chen, S.; Jiang, J.; Chen, F. A bHLH transcription factor regulates iron intake under Fe deficiency in chrysanthemum. Sci. Rep. 2014, 4, 6694. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Zhu, X.; Chen, F.; Gao, H.; Jiang, J.; Chen, S. A chrysanthemum heat shock protein confers tolerance to abiotic stress. Int. J. Mol. Sci. 2014, 15, 5063–5078. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).