In Vivo Delivery Systems for Therapeutic Genome Editing
Abstract
:1. Introduction
2. In Vivo Delivery Systems for Zinc Finger Nucleases (ZFNs) and Their Expression Cassette
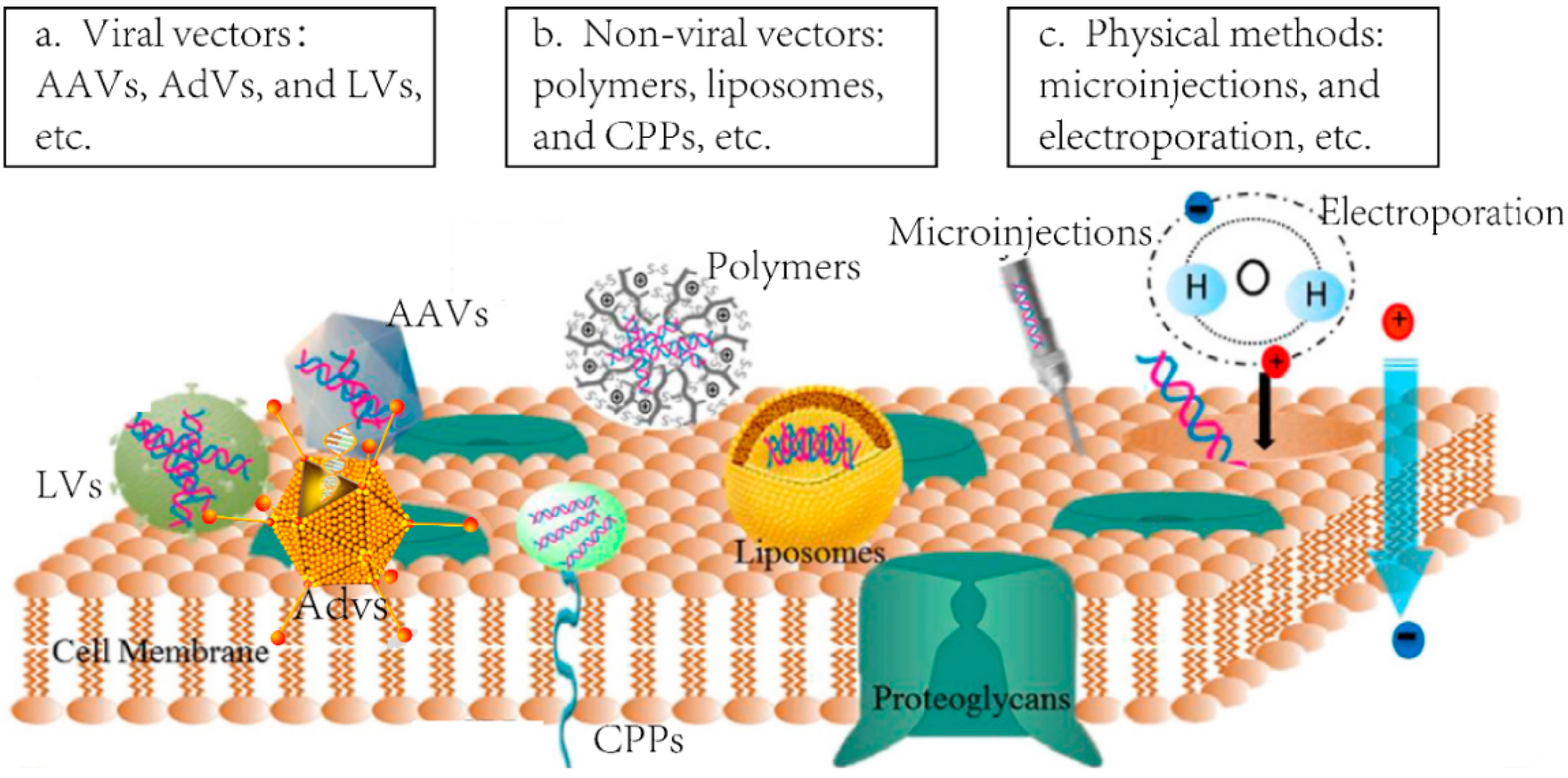
2.1. Viral Delivery
2.1.1. Adenoviral Vectors (AdVs)
2.1.2. Lentiviral Vectors (LVs)
2.1.3. Adeno-Associated Viruses Vectors (AAVs)
2.2. Non-Viral Vectors
2.3. Direct Delivery of ZFN Proteins
2.4. Summary and Prospect
3. In Vivo Delivery Systems for Transcription Activator-Like Effector Nucleases (TALENs) and Their Expression Cassette
3.1. Viral Vectors
3.1.1. Lentiviral Vectors (LVs)
3.1.2. Adenoviral Vectors (AdVs)
3.1.3. Baculoviral Vectors (BVs)
3.2. Non-Viral Vectors
3.2.1. Cationic Polymer-Based Vectors
3.2.2. Conjugates
3.3. Summary and Prospects
4. In Vivo Delivery Systems for CRISPR/Cas9 and Their Expression Cassette
4.1. Viral Vectors
4.2. Non-Viral Vectors
4.2.1. Cationic Lipid-Based Vectors
4.2.2. Cationic Polymer-Based Vectors
4.2.3. Conjugation
4.3. Combined Viral and Non-Viral Delivery
4.4. Summary and Prospects
5. Conclusions and Future Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
Glossary
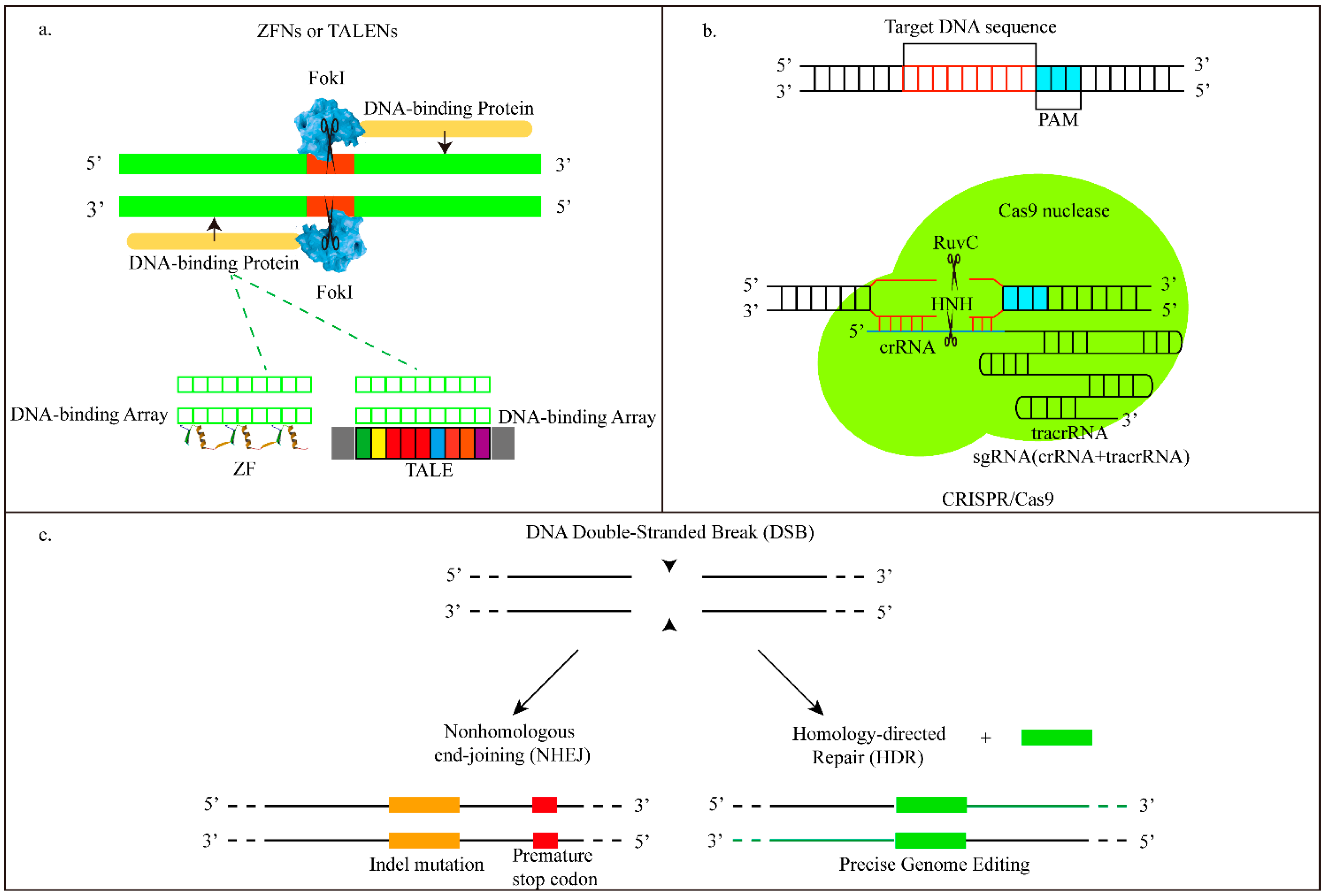
| ZFNs | Zinc-finger nucleases are fusions of the nonspecific DNA-cleavage domain with zinc finger DNA binding domain. The DNA binding domain can be engineered to direct the ZFN to specific desired DNA sequences, and ZFN induce targeted DSBs. |
| TALENs | Transcription activator-like effector nucleases are made by fusing a DNA cleavage domain to a TAL effector DNA binding domain. TAL effectors containing 33~35 amino acid repeat domains can be engineered to recognize base pair. Then TALENs induce targeted DNA DSBs. |
| CRISPR/Cas9 | Clustered regularly interspaced short palindromic repeat- associated nuclease Cas9 is a prokaryotic immune system against foreign DNA in bacteria or archaea. They consist of a guide RNA (tracrRNA-crRNA)-being used for guiding the Cas9 nuclease to the desired DNA sequence, and Cas9 protein. |
| DSB | Double-strand break. A form of DNA damage caused by ZFN, TALEN and CRISPR/Cas9. Both DNA strands will be cleaved. |
| NHEJ | Non-homologous end joining. A DSB repair pathway which can relink broken ends together. It often leads to small insertions and deletions at the broken site. |
| HDR | Homology-directed repair. Another DSB repair pathway by which the donor molecule will be inserted at targeted sites. By HDR, single, multiple transgenes, or single nucleotide substitutions can all be inserted. |
| Indel—insertion/deletion | A molecular biology term. Insert or delete bases in DNA. |
| sgRNA—single-guide RNA | Consists of crRNA and tracrRNA, can direct Cas9-mediated genome editing. |
| PAM | Proto-spacer adjacent motifs on crRNA are particularly required and recognized by Cas9 protein. |
| crRNA | One CRISPR RNA base pair of the guide RNA in CRISPR/Cas9. |
| tracrRNA | Trans-activating chimeric RNA. Another CRISPR RNA base pair in CRISSPR/Cas9. It can promote crRNA processing. |
| Lentiviral Vectors (LVs)—Lentiviruses | The replication-defective retroviruses that are applied to introduce the target gene in vivo or in vitro for genome editing. |
| Adenoviral Vectors (AdVs)—Adenoviruses | The replication-defective viruses belong to the family of Adenoviridae. They are always used to carry the designed gene into targeted tissues or cells in vivo or in vitro for genome editing. |
| Adeno-associated viral Vectors (AAVs)—Adeno-associated viruses | The commonly used delivery vector in genome editing technologies for its potential site-specific integration ability and low immunogenic characteristics. They are icosahedral non-enveloped viruses in Dependovirus genus of Parvoviridae family. |
| Liposomes | Lipid made vesicles which can encapsulate designed DNA or RNA, and carry them into target tissues and cells by fusing to the cell membrane for genome editing in vivo or in vitro. |
| Polymer | Co-polymers made vesicles which can encapsulate both genes and proteins, and form particles with different sizes. They can also be utilized for designed genes or proteins delivery in vivo or in vitro to achieve gene editing therapy. |
| Cell-penetrating peptides (CPP) | Small peptides which can combine with target genes or proteins, and translocate across cell membranes to achieve the delivery and genome editing in vivo or in vitro. |
References
- Cox, D.B.; Platt, R.J.; Zhang, F. Therapeutic genome editing: Prospects and challenges. Nat. Med. 2015, 21, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Perez-Pinera, P.; Ousterout, D.G.; Gersbach, C.A. Advances in targeted genome editing. Curr. Opin. Chem. Biol. 2012, 16, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.C.; Tan, S.; Qiao, G.; Barlow, K.A.; Wang, J.; Xia, D.F.; Meng, X.; Paschon, D.E.; Leung, E.; Hinkley, S.J.; et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011, 29, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; Kim, S.; Kim, J.M.; Kim, J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Wells, D. Gene Therapy Progress and Prospects: Electroporation and other Physical Methods. Nat. Rev. 2004, 11, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nature reviews. Genetics 2003, 4, 346–358. [Google Scholar] [PubMed]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Van Dommelen, S.M.; Vader, P.; Lakhal, S.; Kooijmans, S.A.; van Solinge, W.W.; Wood, M.J.; Schiffelers, R.M. Microvesicles and exosomes: Opportunities for cell-derived membrane vesicles in drug delivery. J. Control. Release 2012, 161, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Vannucci, L.; Lai, M.; Chiuppesi, F.; Ceccherini-Nelli, L.; Pistello, M. Viral vectors: A look back and ahead on gene transfer technology. New Microbiol. 2013, 36, 1–22. [Google Scholar] [PubMed]
- Bessis, N.; GarciaCozar, F.J.; Boissier, M.C. Immune responses to gene therapy vectors: Influence on vector function and effector mechanisms. Gene Ther. 2004, 11, S10–S17. [Google Scholar] [CrossRef] [PubMed]
- Baum, C.; Kustikova, O.; Modlich, U.; Li, Z.; Fehse, B. Mutagenesis and Oncogenesis by Chromosomal Insertion of Gene Transfer Vectors. Hum. Gene Ther. 2006, 17, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Vectors Used in Gene Therapy Clinical Trials. Available online: http://www.wiley.com//legacy/wileychi/genmed/clinical (accessed on July 2015).
- Sun, C.Y.; Shen, S.; Xu, C.F.; Li, H.J.; Liu, Y.; Cao, Z.T.; Yang, X.Z.; Xia, J.X.; Wang, J. Tumor Acidity-Sensitive Polymeric Vector for Active Targeted siRNA Delivery. J. Am. Chem. Soc. 2015, 137, 15217–15224. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Ul Ain, Q.; Chung, J.Y.; Kim, Y.H. Current and future delivery systems for engineered nucleases: ZFN, TALEN and RGEN. J. Control. Release 2015, 205, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Bibikova, M.; Beumer, K.; Trautman, J.K.; Carroll, D. Enhancing Gene Targeting with Designed Zinc Finger Nucleases. Science 2003, 300, 764. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, S.; Huang, X.; Orwig, K.E.; Sheng, Y. Rapid assembly of customized TALENs into multiple delivery systems. PLoS ONE 2013, 8, e80281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urnov, F.D.; Miller, J.C.; Lee, Y.L.; Beausejour, C.M.; Rock, J.M.; Augustus, S.; Jamieson, A.C.; Porteus, M.H.; Gregory, P.D.; Holmes, M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 2005, 435, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.; Dujon, B. Nested chromosomal fragmentation in yeast using the meganuclease I-Sce I: A new method for physical mapping of eukaryotic genomes. Nucleic Acids Res. 1992, 20, 5625–5631. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, B.S.; Stoddard, B.L. Homing endonucleases: Structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001, 29, 3757–3774. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Grizot, S.; Arnould, S.; Duclert, A.; Epinat, J.C.; Chames, P.; Prieto, J.; Redondo, P.; Blanco, F.J.; Bravo, J.; et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006, 34, e149. [Google Scholar] [CrossRef] [PubMed]
- Galetto, R.; Duchateau, P.; Pâques, F. Targeted approaches for gene therapy and the emergence of engineered meganucleases. Expert Opin. Biol. Ther. 2015, 9, 1289–1306. [Google Scholar] [CrossRef] [PubMed]
- Chames, P.; Epinat, J.C.; Guillier, S.; Patin, A.; Lacroix, E.; Pâques, F. In vivo selection of engineered homing endonucleases using double-strand break induced homologous recombination. Nucleic Acids Res. 2005, 33, e178. [Google Scholar] [CrossRef] [PubMed]
- Chapdelaine, P.; Pichavant, C.; Rousseau, J.; Pâques, F.; Tremblay, J.P. Meganucleases can restore the reading frame of a mutated dystrophin. Gene Ther. 2010, 17, 846–858. [Google Scholar] [CrossRef] [PubMed]
- Gouble, A.; Smith, J.; Bruneau, S.; Perez, C.; Guyot, V.; Cabaniols, J.-P.; Leduc, S.; Fiette, L.; Av’e, P.; Micheau, B.; et al. Efficient in toto targeted recombination inmouse liver by meganuclease-induced double-strand break. J. Gene Med. 2006, 8, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Izmiryan, A.; Basmaciogullari, S.; Henry, A.; Paques, F.; Danos, O. Efficient gene targeting mediated by a lentiviral vector-associated meganuclease. Nucleic Acids Res. 2011, 39, 7610–7619. [Google Scholar] [CrossRef] [PubMed]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. 2010, 11, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, J.-S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014, 15, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Blaese, R.M.; Culver, K.W.; Miller, A.D.; Carter, C.S.; Fleisher, T.; Clerici, M.; Shearer, G.; Chang, L.; Chiang, Y.; Tolstoshev, P.; et al. T Lymphocyte-Directed Gene Therapy for ADASCID: Initial Trial Results after 4 Years. Science 1995, 270, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Giacca, M.; Zacchigna, S. Virus-mediated gene delivery for human gene therapy. J. Control. Release 2012, 161, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, T.; Schulz, E.; Ehrhardt, A. Progress and Problems with Viral Vectors for Delivery of TALENs. J. Mol. Genet. Med. 2014, 8, 1–7. [Google Scholar] [CrossRef]
- Campos, S.K.; Barry, M.A. Current Advances and Future Challenges in Adenoviral Vector Biology and Targeting. Curr. Gene Ther. 2007, 7, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Cannon, P.; June, C. Chemokine receptor 5 knockout strategies. Curr. Opin. HIV AIDS 2011, 6, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Kamihira, M. Development of hybrid viral vectors for gene therapy. Biotechnol. Adv. 2013, 31, 208–223. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and Problems with the Use of Viral Vectors for Gene Therapy. Nat. Rev. 2003, 4, 346–359. [Google Scholar] [CrossRef] [PubMed]
- Matrai, J.; Chuah, M.K.; VandenDriessche, T. Recent advances in lentiviral vector development and applications. Mol. Ther. 2010, 18, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, A.; Genovese, P.; Beausejour, C.M.; Colleoni, S.; Lee, Y.L.; Kim, K.A.; Ando, D.; Urnov, F.D.; Galli, C.; Gregory, P.D.; et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007, 25, 1298–1306. [Google Scholar] [CrossRef] [PubMed]
- Apolonia, L.; Waddington, S.N.; Fernandes, C.; Ward, N.J.; Bouma, G.; Blundell, M.P.; Thrasher, A.J.; Collins, M.K.; Philpott, N.J. Stable gene transfer to muscle using non-integrating lentiviral vectors. Mol. Ther. 2007, 15, 1947–1954. [Google Scholar] [CrossRef] [PubMed]
- Gellhaus, K.; Cornu, T.I.; Heilbronn, R.; Cathomen, T. Fate of Recombinant Adeno-Associated Viral Vector Genomes during DNA Double-Strand Break-Induced Gene Targeting in Human Cells. Hum. Gene Ther. 2010, 21, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Ellis, B.L.; Hirsch, M.L.; Porter, S.N.; Samulski, R.J.; Porteus, M.H. Zinc-finger nuclease-mediated gene correction using single AAV vector transduction and enhancement by Food and Drug Administration-approved drugs. Nat. Ther. 2013, 20, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Anguela, X.M.; Sharma, R.; Doyon, Y.; Miller, J.C.; Li, H.; Haurigot, V.; Rohde, M.E.; Wong, S.Y.; Davidson, R.J.; Zhou, S.; et al. Robust ZFN-mediated genome editing in adult hemophilic mice. Blood J. 2013, 122, 3283–3287. [Google Scholar] [CrossRef] [PubMed]
- Händel, E.M.; Gellhaus, K.; Khan, K.; Bednarski, C.; Cornu, T.I.; Müller-Lerch, F.; Kotin, R.M.; Heilbronn, R.; Cathomen, T. Versatile and Efficient Genome Editing in Human Cells by Combining Zinc-Finger Nucleases with Adeno-Associated Viral Vectors. Hum. Gene Ther. 2012, 23, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.H.; Bobis-Wozowicz, S.; Chatterjee, D.; Gellhaus, K.; Pars, K.; Heilbronn, R.; Jacobs, R.; Cathomen, T. The Nontoxic Cell Cycle Modulator Indirubin Augments Transduction of Adeno-Associated Viral Vectors and Zinc-Finger Nuclease-Mediated Gene Targeting. Hum. Gene Ther. 2013, 24, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Michael, J.; Metzger, A.M.-S.; Stoddard, B.L.; Miller, A.D. Single-strand nicks induce homologous recombination with less toxicity than double-strand breaks using an AAV vector template. Nucleic Acids Res. 2011, 39, 926–935. [Google Scholar]
- Hojun, L.; Harigot, V.; Doyon, Y.; Li, J.; Bhagwat, A.; Wong, S.; Anguela, X.; Sharma, R.; Ivanciu, L.; Murphy, S.M.; et al. LBA-5 Phenotypic Correction of a Mouse Model of Hemophilia B by in vivo Genetic Correction of F9 Gene. In Proceedings of the American Society of Hematology Annual Meeting, San Diego, CA, USA, 10–13 December 2010.
- Philip, L.; Felgener, T.R.G.; Holm, M.; Roman, R.; Chen, H.W.; Wenz, M.; Northrop, J.P.; Ringold, G.M.; Dsnielsen, M. Lipofection: A highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA 1987, 84, 7413–7417. [Google Scholar]
- Almofti, M.R.; Harashima, H.; Shinohara, Y.; Almofti, A.; Baba, Y.; Kiwada, H. Cationic liposome-mediated gene delivery: Biophysical study and mechanism of internalization. Arch. Biochem. Biophys. 2003, 410, 246–253. [Google Scholar] [CrossRef]
- Perez, E.E.; Wang, J.B.; Miller, J.C.; Jouvenot, Y.; Kim, K.A.; Liu, O.; Wang, N.; Lee, G.; Bartsevich, V.V.; Lee, Y.; et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biothechnol. 2008, 26, 808–817. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.W.; Baginski, T.K.; Reilly, D.E. Reilly, Enhancement of DNA Uptake in FUT8-Deleted CHO Cells for Transient Production of Afucosylated Antibodies. Biotechnol. Bioeng. 2010, 106, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Gaj, T.; Guo, J.; Kato, Y.; Sirk, S.J.; Barbas, C.F., 3rd. Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat. Methods 2012, 9, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J.; Kim, J. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, S.; Kim, Y.H.; Kim, H. Stability of Zinc Finger Nuclease Protein Is Enhanced by the Proteasome Inhibitor MG132. PLoS ONE 2013, 8, e54282. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.I.; Kim, H.; Ramakrishna, S. Recent developments and clinical studies utilizing engineered zinc finger nuclease technology. Cell. Mol. Life Sci. 2015, 72, 3819–3830. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Haurigot, V.; Doyon, Y.; Li, T.; Wong, S.Y.; Bhagwat, A.S.; Malani, N.; Anguela, X.M.; Sharma, R.; Ivanciu, L.; et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 2011, 475, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Mussolino, C.; Morbitzer, R.; Lütge, F.; Dannemann, N.; Lahaye, T.; Cathomen, T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011, 39, 9283–9293. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Lau, C.H.; Goh, S.L.; Liang, Q.; Chen, C.; Du, S.; Phang, R.Z.; Tay, F.C.; Tan, W.K.; Li, Z.; et al. Baculoviral transduction facilitates TALEN-mediated targeted transgene integration and Cre/LoxP cassette exchange in human-induced pluripotent stem cells. Nucleic Acids Res. 2013, 41, e180. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Yang, Y. Targeted Genome Editing Tools for Disease Modeling and Gene Therapy. Curr. Gene Ther. 2014, 14, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.E.; Hakim, C.H.; Ousterout, D.G.; Thakore, P.I.; Moreb, E.A.; Rivera, R.M.C.; Zhang, F.; Madhavan, S.; Duan, D.; Ousterout, D.G.; et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 2016, 351, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Holkers, M.; de Vries, A.A.F.; Gonçalves, M.A.F.V. Nonspaced inverted DNA repeats are preferential targets for homology-directed gene repair in mammalian cells. Nucleic Acids Res. 2012, 40, 1984–1999. [Google Scholar] [CrossRef] [PubMed]
- Holkers, M.; Maggio, I.; Liu, J.; Janssen, J.M.; Miselli, F.; Mussolino, C.; Recchia, A.; Cathomen, T.; Gonçalves, M.A. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2012, 41, e63. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Ding, W.; Zhu, D.; Yu, L.; Jiang, X.; Wang, X.; Zhang, C.; Wang, L.; Ji, T.; Liu, D.; et al. TALEN-mediated targeting of HPV oncogenes ameliorates HPV-related cervical malignancy. J. Clin. Investig. 2015, 125, 425–537. [Google Scholar] [CrossRef] [PubMed]
- Ru, R.; Yao, Y.; Yu, S.; Yin, B.; Xu, W.; Zhao, S.; Qin, L.; Chen, X. Targeted genome engineering in human induced pluripotent stem cells by penetrating TALENs. Cell Regen. 2013, 2, 5. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Gaj, T.; Patterson, J.T.; Sirk, S.J.; Barbas, C.F. Cell-penetrating peptide-mediated delivery of TALEN proteins via bioconjugation for genome engineering. PLoS ONE 2014, 9, e85755. [Google Scholar] [CrossRef] [PubMed]
- Wiedenheft, B.; Sternberg, S.H.; Doudna, J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nat. Rev. 2012, 482, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Peter, C.; Fineran, E.C. Memory of viral infections by CRISPR-Cas adaptive immune systems: Acquisition of new information. Virology 2012, 434, 202–209. [Google Scholar]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance against Viruses in Prokaryotes. Sci. Rep. 2007, 315, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the Immune System of Bacteria and Archaea. Science 2010, 327, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Aravind, L.; Grishin, N.V.; Rogozin, I.B.; Koonin, E.V. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002, 30, 482–496. [Google Scholar] [CrossRef] [PubMed]
- Heidenreich, M.; Zhang, F. Applications of CRISPR-Cas system in neuroscience. Nat. Rev. Neurosci. 2015, 17, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Swiech, L.; Heidenreich, M.; Banerjee, A.; Habib, N.; Li, Y.; Trombetta, J.; Sur, M.; Zhang, F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 2015, 33, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Tabebordbar, M.; Zhu, K.; Cheng, J.K.; Chew, W.L.; Widrick, J.J.; Yan, W.X.; Maesner, C.; Wu, E.Y.; Xiao, R.; Ran, F.A.; et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016, 351, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; Amoasii, L.; Mireault, A.A.; McAnally, J.R.; Li, H.; Sanchez-Ortiz, E.; Bhattacharyya, S.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 2016, 351, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Yue, Y.; Liu, M.; Ghosh, A.; Engelhardt, J.F.; Chamberlain, J.S.; Duan, D. Efficient in vivo gene expression by trans-splicing adeno-associated viral vectors. Nat. Biotechnol. 2005, 23, 1435–1439. [Google Scholar] [CrossRef] [PubMed]
- Murlidharan, G.; Samulski, R.J.; Asokan, A. Biology of adeno-associated viral vectors in the central nervous system. Front. Mol. Neurosci. 2014, 7, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Platt, R.J.; Chen, S.; Zhou, Y.; Yim, M.J.; Swiech, L.; Kempton, H.R.; Dahlman, J.E.; Parnas, O.; Eisenhaure, T.M.; Jovanovic, M.; et al. CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling. Cell 2014, 159, 440–455. [Google Scholar] [CrossRef] [PubMed]
- Senis, E.; Fatouros, C.; Grosse, S.; Wiedtke, E.; Niopek, D.; Mueller, A.K.; Borner, K.; Grimm, D. CRISPR/Cas9-mediated genome engineering: An adeno-associated viral (AAV) vector toolbox. Biotechnol. J. 2014, 9, 1402–1412. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, E.M.; Kornepati, A.V.; Mefferd, A.L.; Marshall, J.B.; Tsai, K.; Bogerd, H.P.; Cullen, B.R. Optimization of a multiplex CRISPR/Cas system for use as an antiviral therapeutic. Methods 2015, 91, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.; Peng, J.; Yan, Y.; Cao, P.; Wang, J.; Qiu, C.; Tang, L.; Liu, D.; Tang, L.; Jin, J.; et al. Efficient gene editing in adult mouse livers via adenoviral delivery of CRISPR/Cas9. FEBS Lett. 2014, 588, 3954–3958. [Google Scholar] [CrossRef] [PubMed]
- Zuris, J.A.; Thompson, D.B.; Shu, Y.; Guilinger, J.P.; Bessen, J.L.; Hu, J.H.; Maeder, M.L.; Joung, J.K.; Chen, Z.Y.; Liu, D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015, 33, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Wightman, L.; Kircheis, R.; Rössler, V.; Carotta, S.; Ruzicka, R.; Kursa, M.; Wagner, E. Different behavior of branched and linear polyethylenimine for gene delivery in vitro and in vivo. J. Gene Med. 2011, 3, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Zuckermann, M.; Hovestadt, V.; Knobbe-Thomsen, C.B.; Zapatka, M.; Northcott, P.A.; Schramm, K.; Belic, J.; Jones, D.T.; Tschida, B.; Moriarity, B.; et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat. Commun. 2015, 6, 7391–7400. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; He, Z.Y.; Wei, X.W.; Gao, G.P.; Wei, Y.Q. Challenges in CRISPR/CAS9 Delivery: Potential Roles of Nonviral Vectors. Hum. Gene Ther. 2015, 26, 452–462. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, R.D.; Sill, M.W.; Davidson, S.A.; Muller, C.Y.; Bender, D.P.; DeBernardo, R.L.; Behbakht, K.; Huh, W.K. A phase II trial of intraperitoneal EGEN-001, an IL-12 plasmid formulated with PEG–PEI-cholesterol lipopolymer in the treatment of persistent of recurrent epithelial ovarian, fallopian tube or primary peritoneal cancer: A gynecologic oncology group study. Gynecol. Oncol. 2014, 3, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, S.; Kwaku Dad, A.B.; Beloor, J.; Gopalappa, R.; Lee, S.K.; Kim, H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014, 24, 1020–1027. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Song, C.Q.; Dorkin, J.R.; Zhu, L.J.; Li, Y.; Wu, Q.; Park, A.; Yang, J.; Suresh, S.; Bizhanova, A.; et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016, 34, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Findlay, G.M.; Boyle, E.A.; Hause, R.J.; Klein, J.C.; Shendure, J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature 2014, 513, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Xue, W.; Chen, S.; Bogorad, R.L.; Benedetti, E.; Grompe, M.; Koteliansky, V.; Sharp, P.A.; Jacks, T.; Anderson, D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014, 32, 551–553. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Chen, S.; Yin, H.; Tammela, T.; Papagiannakopoulos, T.; Joshi, N.S.; Cai, W.; Yang, G.; Bronson, R.; Crowley, D.G. CRISPR-mediated directmutation of cancer genes in themouse liver. Nature 2014, 514, 380–396. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.I.; Suresh, B.; Kim, H.; Ramakrishna, S. CRISPR/Cas9 system as an innovative genetic engineering tool: Enhancements in sequence specificity and delivery methods. Biochim. Biophys. Acta 2015, 1856, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Rieder, F.; Steininger, C. Cytomegalovirus vaccine: Phase II clinical trial results. Clin. Microbiol. Infect. 2013, 20, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Lee, H.J.; Carroll, D. Genome editing with modularly assembled zinc-finger nucleases. Nat. Methods 2010, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kweon, J.; Kim, A.; Chon, J.K.; Yoo, J.Y.; Kim, H.J.; Kim, S.; Lee, C.; Jeong, E.; Chung, E.; et al. A library of TAL effector nucleases spanning the human genome. Nat. Biotechnol. 2013, 31, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Cornu, T.I.; Thibodeau-Beganny, S.; Guhl, E.; Alwin, S.; Eichtinger, M.; Joung, J.K.; Cathomen, T. DNA-binding Specificity Is a Major Determinant of the Activity and Toxicity of Zinc-finger Nucleases. Mol. Ther. 2008, 16, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Maggio, I.; Holkers, M.; Liu, J.; Janssen, J.M.; Chen, X.; Gonçalves, M.A.F.V. Adenoviral vector delivery of RNA-guided CRISPR/Cas9 nuclease complexes induces targeted mutagenesis in a diverse array of human cells. Sci. Rep. 2014, 4, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science 2014, 343, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wei, J.J.; Sabatini, D.M.; Lander, E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 2014, 343, 80–86. [Google Scholar] [CrossRef] [PubMed]
- McNeer, N.A.; Anandalingam, K.; Fields, R.J.; Caputo, C.; Kopic, S.; Gupta, A.; Quijano, E.; Polikoff, L.; Kong, Y.; Bahal, R.; et al. Nanoparticles that deliver triplex-forming peptide nucleic acid molecules correct F508del CFTR in airway epithelium. Nat. Commun. 2015, 6, 6952. [Google Scholar] [CrossRef] [PubMed]
- Tasciotti, E.; Liu, X.; Bhavane, R.; Plant, K.; Leonard, A.D.; Price, B.K.; Cheng, M.M.; Decuzzi, P.; Tour, J.M.; Robertson, F.; et al. Mesoporous silicon particles as a multistage delivery system for imaging and therapeutic applications. Nat. Nanotechnol. 2008, 3, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.H.; Yu, S.Y. Cationic nanoemulsions as non-viral vectors for plasmid DNA delivery. Colloids Surf. B Biointerfaces 2010, 79, 509–515. [Google Scholar] [CrossRef] [PubMed]
- He, Z.Y.; Wei, X.W.; Luo, M.; Luo, S.T.; Yang, Y.; Yu, Y.Y.; Chen, Y.; Ma, C.C.; Liang, X.; Guo, F.C. Folate-linked lipoplexes for short hairpin RNA targeting claudin-3 delivery in ovarian cancer xenografts. J. Control. Release 2013, 172, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; Kim, S.; Kim, Y.; Kweon, J.; Kim, H.S.; Bae, S.; Kim, J. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014, 24, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Francisco Martín, K.B.; Cobo, M.; Muñoz, P.; Anderson, P.; Toscano, M.G. New Vectors for Stable and Safe Gene Modification. Gene Therapy—Developments and Future Perspectives; InTech: Rijeka, Croatia, 2011; pp. 1–29. ISBN 978-953-307-617-1. [Google Scholar]
- Klein, T.; Loschberger, A.; Proppert, S.; Wolter, S.; van de Linde, S.; Sauer, M. Live-cell dSTORM with SNAP-tag fusion proteins. Nat. Methods 2011, 8, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Tebas, P.; Stein, D.; Tang, W.W.; Frank, I.; Wang, S.Q.; Lee, G.; Spratt, S.K.; Surosky, R.T.; Giedlin, M.A.; Nichol, G.; et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014, 370, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Lee, H.J.; Kim, H.; Cho, S.W.; Kim, J.S. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009, 19, 1279–1288. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kweon, J.; Kim, J.S. TALENs and ZFNs are associated with different mutation signatures. Nat. Methods 2013, 10, 185. [Google Scholar] [CrossRef] [PubMed]
- Burnett, J.C.; Zaia, J.A.; Rossi, J.J. Creating genetic resistance to HIV. Curr. Opin. Immunol. 2012, 24, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Qasim, W.; Amrolia, P.J.; Samarasinghe, S.; Ghorashian, S.; Zhan, H.; Stafford, S.; Butler, K.; Ahsan, G.; Gilmour, K.; Adams, S.; et al. First Clinical Application of Talen Engineered Universal CAR19 T Cells in B-ALL. Blood 2015, 126, 2046. [Google Scholar]
| Typical Delivery Systems | Assessment | Genome Editing Nuclease | Clinical Trials | |||||
|---|---|---|---|---|---|---|---|---|
| Advantages | Disadvantages | Phase | Status | Clinical Trials. Gov Identifier | Reference | |||
| AAVs | High efficiency | Low packaging capacity, cost high | ZFNs, CRISPR/Cas9 | – | – | – | [59,73,74,104] | |
| AdVs | Low off-target mutagenesis | Immunoreactivity, high cost | ZFNs | I | Completed | NCT01044654 | [49,105,106] | |
| II | Completed | NCT01252641 | ||||||
| I | Completed | NCT00842634 | ||||||
| HCAdVs | High packaging capacity | Cell-specific targeting is difficult to achieve | TALENs | – | – | – | [32] | |
| CPP, e.g., TAT-TALEN proteins; CPP-Cas9 proteins | Low off-target mutagenesis | Immunoreactivity | TALENs, CRISPR/Cas9 | – | – | – | [63,64,86] | |
| Candidates for delivering plasmids of nucleases | DOTAP-cholesterol | Easy to produce, large packaging capacity | Large particle size, low targeting efficiency, toxic | – | I/II | Active | NCT01455389 | [8] |
| PEI | Easy to produce, large packaging capacity | Low targeting efficiency, toxic | – | II | Active | NCT00595088 | ||
| PEG-PEI-Cholesterol | Easy to produce, large packaging capacity with small particle size, low toxic | Low targeting efficiency | – | II | Active | NCT01118052 | ||
| Genome Editing Nucleases | DNA Targeting Specificity Determinant | Endonuclease | Average Mutation Rate | Off-Target Rate | Success Rate | Size | Cytotoxicity |
|---|---|---|---|---|---|---|---|
| ZFNs | Zinc-finger proteins | FokI | 10% | High | ~24% | ~1 kb × 2 | Variable~high |
| TALENs | Transcription activator-like effectors | FokI | 20% | Low | ~99% | ~3 kb × 2 | Low |
| CRISPR/Cas9 | crRNA or sgRNA | Cas9 | 20% | Variable | ~90% | 4.2 kb (SpCas9) + 0.1 kb (sgRNA) | Low |
| Genome Editing Nucleases | Clinical Trials | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Condition | Intervention | Target | Delivery Vector | Cell Transplantation | Company | Phase | Status | Clinical Trials. Gov Identifier | Reference | |
| ZFNs | HIV Infection | Genetic: SB-728-T | CCR5 DNA | AdVs or direct delivery | Autologous CD4+ T cells | Sangamo Biosciences | I | Completed | NCT01044654 | [49,106,109] |
| II | Completed | NCT01252641 | ||||||||
| I | Completed | NCT00842634 | ||||||||
| TALENs | Leukemia | Malignant blood cells | Genes in immune cells | N/A | Chimeric antigen receptor (CAR) 19 T cells | Great Ormond Street Hospital | N/A | N/A | N/A | [110] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Li, F.; Dang, L.; Liang, C.; Wang, C.; He, B.; Liu, J.; Li, D.; Wu, X.; Xu, X.; et al. In Vivo Delivery Systems for Therapeutic Genome Editing. Int. J. Mol. Sci. 2016, 17, 626. https://doi.org/10.3390/ijms17050626
Wang L, Li F, Dang L, Liang C, Wang C, He B, Liu J, Li D, Wu X, Xu X, et al. In Vivo Delivery Systems for Therapeutic Genome Editing. International Journal of Molecular Sciences. 2016; 17(5):626. https://doi.org/10.3390/ijms17050626
Chicago/Turabian StyleWang, Luyao, Fangfei Li, Lei Dang, Chao Liang, Chao Wang, Bing He, Jin Liu, Defang Li, Xiaohao Wu, Xuegong Xu, and et al. 2016. "In Vivo Delivery Systems for Therapeutic Genome Editing" International Journal of Molecular Sciences 17, no. 5: 626. https://doi.org/10.3390/ijms17050626