A Proteomic Approach for the Identification of Up-Regulated Proteins Involved in the Metabolic Process of the Leiomyoma
Abstract
:1. Introduction
2. Result
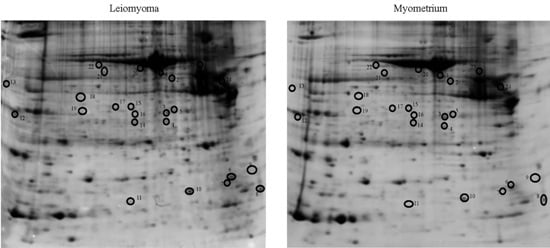
2.1. Proteomic Studies
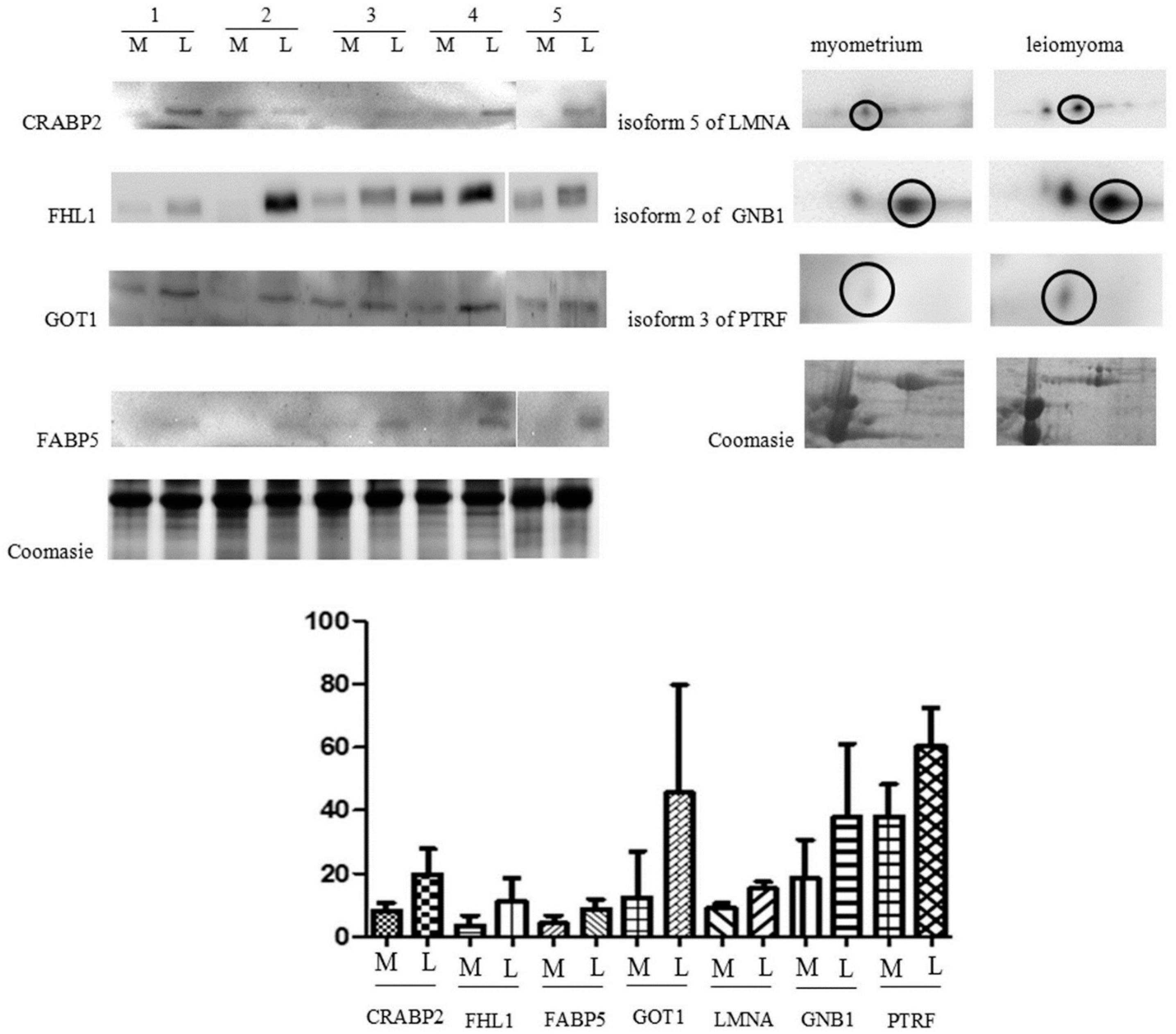
2.2. Immunochemical Study of Proteins Expression
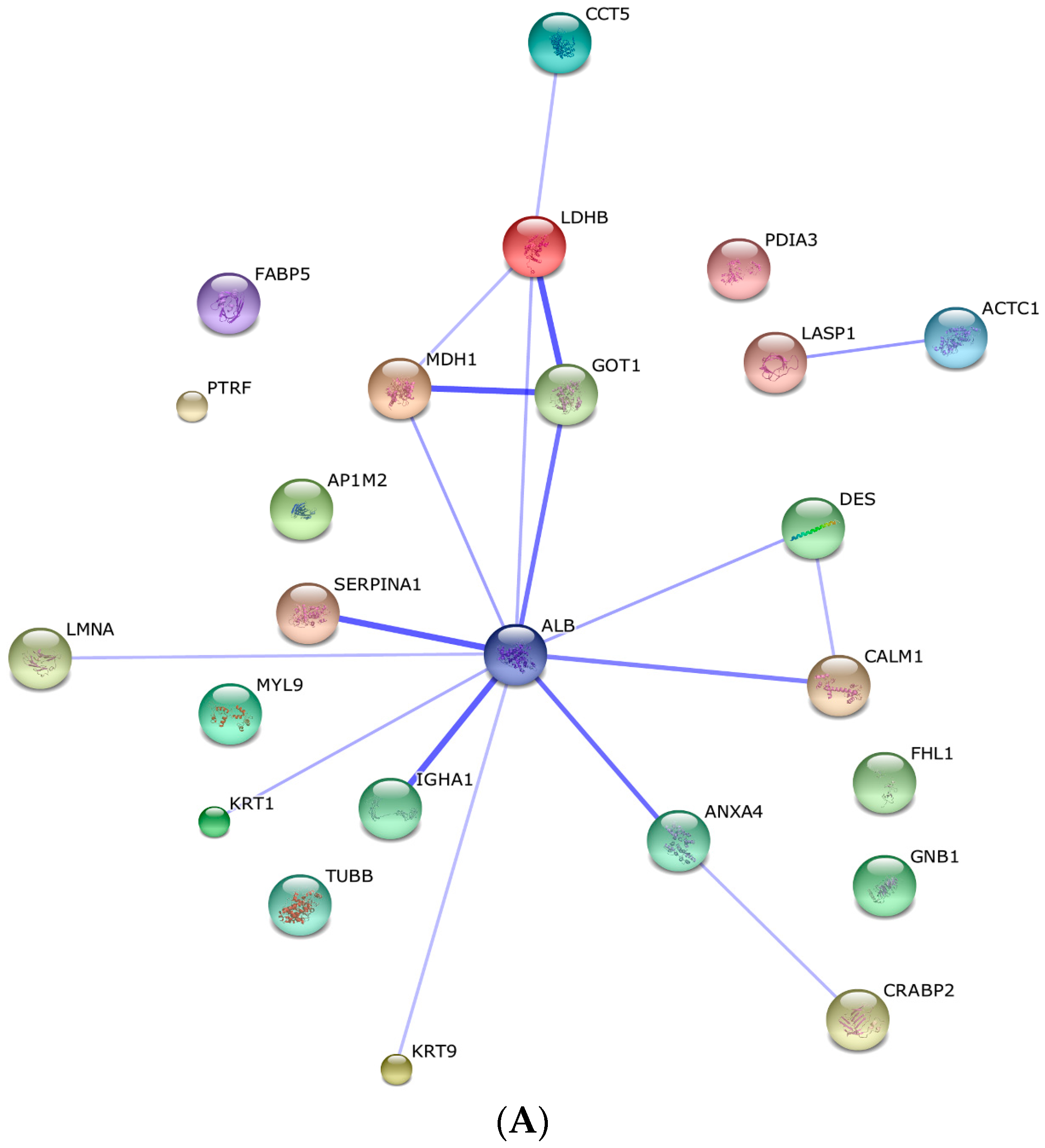
2.3. Protein–Protein Interaction and Co-Expression Study
2.4. Functional Analysis of the Leiomyoma Proteome
3. Discussion
4. Experimental Section
4.1. Tissue Samples
4.2. Two-Dimensional Gel Electrophoresis
4.3. Quantification of Spot Levels
4.4. Trypsin Digestion
4.5. MS Analysis
4.6. Protein–Protein Interaction, Co-Expression and Functional Analysis
4.7. Western Blot Analysis
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Okolo, S. Incidence, aetiology and epidemiology of uterine fibroids. Best Pract. Res. Clin. Obstet. Gynaecol. 2008, 22, 571–588. [Google Scholar] [CrossRef] [PubMed]
- Haney, A.F. Clinical decision making regarding leiomyomata: What we need in the next millennium. Environ. Health Perspect. 2000, 108, 835–839. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Zhu, X.; Dong, K.; Lin, Y.; Hu, Y.; Zhu, C. Reduced expression of 14-3-3 γ in uterine leiomyoma as identified by proteomics. Fertil. Steril. 2008, 90, 1892–1898. [Google Scholar] [CrossRef] [PubMed]
- Flake, G.P.; Andersen, J.; Dixon, D. Etiology and pathogenesis of uterine leiomyomas: A review. Environ. Health Perspect. 2003, 8, 1037–1054. [Google Scholar] [CrossRef]
- Takeda, T.; Sakata, M.; Isobe, A.; Miyake, A.; Nishimoto, F.; Ota, Y.; Kamiura, S.; Kimura, T. Relationship between metabolic syndrome and uterine leiomyomas: A case-control study. Gynecol. Obstet. Investig. 2008, 66, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Takeda, T.; Tsuiji, K.; Kondo, A.; Kitamura, M.; Wong, T.F.; Yaegashi, N. The antidiabetic drug metformin inhibits uterine leiomyoma cell proliferation via an AMP-activated protein kinase signaling pathway. Gynecol. Endocrinol. 2013, 29, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Ciavattini, A.; di Giuseppe, J.; Stortoni, P.; Montik, N.; Giannubilo, S.R.; Litta, P.; Islam, S.; Tranquilli, A.L.; Reis, F.M.; Ciarmela, P. Uterine fibroids: Pathogenesis and interactions with endometrium and endomyometrial junction. Obstet. Gynecol. Int. 2013. [Google Scholar] [CrossRef] [PubMed]
- Arslan, A.A.; Gold, L.I.; Mittal, K.; Suen, T.C.; Belitskaya-Levy, I.; Tang, M.S.; Toniolo, P. Gene expression studies provide clues to the pathogenesis of uterine leiomyoma: New evidence and a systematic review. Hum. Reprod. 2005, 20, 852–863. [Google Scholar] [CrossRef] [PubMed]
- Turunen, M.; Spaeth, J.M.; Keskitalo, S.; Park, M.J.; Kivioja, T.; Clark, A.D.; Mäkinen, N.; Gao, F.; Palin, K.; Nurkkala, H.; et al. Uterine leiomyoma-linked MED12 mutations disrupt mediator-associated CDK activity. Cell Rep. 2014, 7, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Morgan, E.; Kannan-Thulasiraman, P.; Noy, N. Involvement of fatty acid binding protein 5 and PPARβ/δ in prostate cancer cell growth. PPAR Res. 2010. [Google Scholar] [CrossRef] [PubMed]
- Schug, T.T.; Berry, D.C.; Toshkov, I.A.; Cheng, L.; Nikitin, A.Y.; Noy, N. Overcoming retinoic acid-resistance of mammary carcinomas by diverting retinoic acid from PPARβ/δ to RAR. Proc. Natl. Acad. Sci. USA 2008, 105, 7546–7551. [Google Scholar] [CrossRef] [PubMed]
- Tordjman, J.; Leroyer, S.; Chauvet, G.; Quette, J.; Chauvet, C.; Tomkiewicz, C.; Chapron, C.; Barouki, R.; Forest, C.; Aggerbeck, M.; et al. Cytosolic aspartate aminotransferase, a new partner in adipocyte glyceroneogenesis and an atypical target of thiazolidinedione. J. Biol. Chem. 2007, 282, 23591–23602. [Google Scholar] [CrossRef] [PubMed]
- Ura, B.; Scrimin, F.; Zanconati, F.; Arrigoni, G.; Monasta, L.; Romano, A.; Banco, R.; Zweyer, M.; Milani, D.; Ricci, G. Two-dimensional gel electrophoresis analysis of the leiomyoma interstitial fluid reveals altered protein expression with a possible involvement in pathogenesis. Oncol. Rep. 2015, 33, 2219–2226. [Google Scholar] [CrossRef] [PubMed]
- Lemeer, S.; Gholami, A.M.; Wu, Z.; Kuster, B. Quantitative Proteome Profiling of human Myoma and Myometrium tissue reveals kinase expression signatures with potential for therapeutic intervention. Proteomics 2015, 15, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Okada, S.; Jørgensen, J.O.; Kopchick, J.J. Novel serum protein biomarkers indicative of growth hormone doping in healthy human subjects. Proteomics 2011, 17, 3565–3571. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, E.B.; Colowick, S.P. The Role of glycolysis in the growth of tumor cells.3. Lactic dehydrogenase as the site of action of oxamate on the growth of cultured cells. J. Biol. Chem. 1965, 240, 2786–2790. [Google Scholar] [PubMed]
- Thornburg, J.M.; Nelson, K.K.; Clem, B.F.; Lane, A.N.; Arumugam, S.; Simmons, A.; Eaton, J.W.; Telang, S.; Chesney, J. Targeting aspartate aminotransferase in breast cancer. Breast Cancer Res. 2008. [Google Scholar] [CrossRef] [PubMed]
- Zaitseva, M.; Vollenhoven, B.J.; Rogers, P.A. Retinoic acid pathway genes show significantly altered expression in uterine fibroids when compared with normal myometrium. Mol. Hum. Reprod. 2007, 13, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Connolly, R.M.; Nguyen, N.K.; Sukumar, S. Molecular pathways: Current role and future directions of the retinoic acid pathway in cancer prevention and treatment. Clin. Cancer Res. 2013, 19, 1651–1659. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.M.; Wang, P.; Zhu, B.T. Characterization of the estradiol-binding site structure of human protein disulfide isomerase (PDI). PLoS ONE 2011. [Google Scholar] [CrossRef] [PubMed]
- Spiess, C.; Meyer, A.S.; Reissmann, S.; Frydman, J. Mechanism of the eukaryotic chaperonin: Protein folding in the chamber of secrets. Trends Cell Biol. 2004, 14, 598–604. [Google Scholar] [CrossRef] [PubMed]
- Yaffe, M.B.; Farr, G.W.; Miklos, D.; Horwich, A.L.; Sternlicht, M.L.; Sternlicht, H. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature 1992, 358, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Cowling, B.S.; McGrath, M.J.; Nguyen, M.A.; Cottle, D.L.; Kee, A.J.; Brown, S.; Schessl, J.; Zou, Y.; Joya, J.; Bönnemann, C.G.; et al. Identification of FHL1 as a regulator of skeletal muscle mass: Implications for human myopathy. J. Cell Biol. 2008, 183, 1033–1048. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, R.; Yagi, S.; Ohgane, J.; Yamagata, Y.; Asada, H.; Tamura, I.; Sugino, N.; Shiota, K. Disease-dependent differently methylated regions (D-DMRs) of DNA are enriched on the X chromosome in uterine leiomyoma. J. Reprod. Dev. 2011, 57, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Crabtree, J.S.; Jelinsky, S.A.; Harris, H.A.; Choe, S.E.; Cotreau, M.M.; Kimberland, M.L.; Wilson, E.; Saraf, K.A.; Liu, W.; McCampbell, A.S.; et al. Comparison of human and rat uterine leiomyomata: Identification of a dysregulated mammalian target of rapamycin pathway. Cancer Res. 2009, 69, 6171–6178. [Google Scholar] [CrossRef] [PubMed]
- Mogami, T.; Yokota, N.; Asai-Sato, M.; Yamada, R.; Koizume, S.; Sakuma, Y.; Yoshihara, M.; Nakamura, Y.; Takano, Y.; Hirahara, F.; et al. Annexin A4 is involved in proliferation, chemo-resistance and migration and invasion in ovarian clear cell adenocarcinoma cells. PLoS ONE 2013. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Enomoto, T.; Serada, S.; Ueda, Y.; Takahashi, T.; Ripley, B.; Miyatake, T.; Fujita, M.; Lee, C.M.; Morimoto, K.; et al. Enhanced expression of Annexin A4 in clear cell carcinoma of the ovary and its association with chemoresistance to carboplatin. Int. J. Cancer 2009, 125, 2316–2322. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Wang, J.; Hou, L.; Li, J.; Chen, G.; Jing, B.; Zhang, X.; Yang, Z. Annexin A1, A2, A4 and A5 play important roles in breast cancer, pancreatic cancer and laryngeal carcinoma, alone and/or synergistically. Oncol. Lett. 2013, 5, 107–112. [Google Scholar] [PubMed]
- Ura, B.; Scrimin, F.; Monasta, L.; Radillo, O.; Ricci, G. Association between up-regulated expression proteins and circulating steroidal hormones in leiomyoma. Med. Hypotheses 2015. [Google Scholar] [CrossRef] [PubMed]
- Manavathi, B.; Acconcia, F.; Rayala, S.K.; Kumar, R. An inherent role of microtubule network in the action of nuclear receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 15981–15986. [Google Scholar] [CrossRef] [PubMed]
- Salama, S.A.; Kamel, M.W.; Botting, S.; Salih, S.M.; Borahay, M.A.; Hamed, A.A.; Kilic, G.S.; Saeed, M.; Williams, M.Y.; Diaz-Arrastia, C.R. Catechol-o-methyltransferase expression and 2-methoxyestradiol affect microtubule dynamics and modify steroid receptor signaling in leiomyoma cells. PLoS ONE 2009. [Google Scholar] [CrossRef] [PubMed]
- Pixley, R.A.; Espinola, R.G.; Ghebrehiwet, B.; Joseph, K.; Kao, A.; Bdeir, K.; Cines, D.B.; Colman, R.W. Interaction of high-molecular-weight kininogen with endothelial cell binding proteins suPAR, gC1qR and cytokeratin 1 determined by surface plasmon resonance (BiaCore). Thromb. Haemost. 2011, 105, 1053–1059. [Google Scholar] [CrossRef] [PubMed]
- Nierth-Simpson, E.N.; Martin, M.M.; Chiang, T.C.; Melnik, L.I.; Rhodes, L.V.; Muir, S.E.; Burow, M.E.; McLachlan, J.A. Human uterine smooth muscle and leiomyoma cells differ in their rapid 17-β-estradiol signaling: Implications for proliferation. Endocrinology 2009, 150, 2436–2445. [Google Scholar] [CrossRef] [PubMed]
- Reichow, S.L.; Clemens, D.M.; Freites, J.A.; Németh-Cahalan, K.L.; Heyden, M.; Tobias, D.J.; Hall, J.E.; Gonen, T. Allosteric mechanism of water-channel gating by Ca2+-calmodulin. Nat. Struct. Mol. Biol. 2013, 20, 1085–1092. [Google Scholar] [CrossRef] [PubMed]
- Berggård, T.; Arrigoni, G.; Olsson, O.; Fex, M.; Linse, S.; James, P. 140 mouse brain proteins identified by Ca2+-calmodulin affinity chromatography and tandem mass spectrometry. J. Proteome Res. 2006, 5, 669–687. [Google Scholar] [CrossRef] [PubMed]
- Kahl, C.R.; Means, A.R. Regulation of cell cycle progression by calcium/calmodulin-dependent pathways. Endocr Rev. 2003, 24, 719–736. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.C.; Chi, N.H.; Chou, N.K.; Huang, Y.Y.; Chung, T.W.; Chang, Y.L.; Liu, H.C.; Shieh, M.J.; Wang, S.S. The influence of rat mesenchymal stem cell CD44 surface markers on cell growth, fibronectin expression, and cardiomyogenic differentiation on silk fibroin-Hyaluronic acid cardiac patches. Biomaterials 2010, 31, 854–862. [Google Scholar] [CrossRef] [PubMed]
- Rogers, R.; Norian, J.; Malik, M.; Christman, G.; Abu-Asab, M.; Chen, F.; Korecki, C.; Iatridis, J.; Catherino, W.H.; Tuan, R.S.; et al. Mechanical homeostasis is altered in uterine leiomyoma. Am. J. Obstet. Gynecol. 2008, 198, 474.e1–474.e11. [Google Scholar] [CrossRef] [PubMed]
- Ohno, H.; Tomemori, T.; Nakatsu, F.; Okazaki, Y.; Aguilar, R.C.; Foelsch, H.; Mellman, I.; Saito, T.; Shirasawa, T.; Bonifacino, J.S. Mu1B, a novel adaptor medium chain expressed in polarized epithelial cells. FEBS Lett. 1999, 449, 215–220. [Google Scholar] [CrossRef]
- Park, J.E.; Tan, H.S.; Datta, A.; Lai, R.C.; Zhang, H.; Meng, W.; Lim, S.K.; Sze, S.K. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol. Cell. Proteom. 2010, 9, 1085–1099. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, T.G.; Kammerer, U.; Schulze, E.; Schindler, D.; Honig, A.; Zimmer, M.; Butt, E. Silencing of LASP-1 influences zyxin localization, inhibits proliferation and reduces migration in breast cancer cells. Exp. Cell Res. 2006, 312, 974–982. [Google Scholar] [CrossRef] [PubMed]
- Millioni, R.; Iori, E.; Puricelli, L.; Arrigoni, G.; Vedovato, M.; Trevisan, R.; James, P.; Tiengo, A.; Tessari, P. Abnormal cytoskeletal protein expression in cultured skin fibroblasts from type 1 diabetes mellitus patients with nephropathy: A proteomic approach. Proteom. Clin. Appl. 2008, 2, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Carcoforo, P.; Ura, B.; Mischiati, C.; Squerzanti, M.; Lanzara, V.; Cervellati, C.; Calza, R.; de Laureto, P.P.; Frare, E.; Portinari, M.; et al. Comparative proteomic analysis of ductal breast carcinoma demonstrates an altered expression of chaperonins and cytoskeletal proteins. Mol. Med. Rep. 2013, 7, 1700–1704. [Google Scholar] [PubMed]
- Chillà, A.; Magherini, F.; Margheri, F.; Laurenzana, A.; Gamberi, T.; Bini, L.; Bianchi, L.; Danza, G.; Mazzanti, B.; Serratì, S.; et al. Proteomic identification of VEGF-dependent protein enrichment to membrane caveolar-raft microdomains in endothelial progenitor cells. Mol. Cell. Proteom. 2013, 12, 1926–1938. [Google Scholar] [CrossRef] [PubMed]
- Mischiati, C.; Ura, B.; Roncoroni, L.; Elli, L.; Cervellati, C.; Squerzanti, M.; Conte, D.; Doneda, L.; de Laureto, P.; de Franceschi, G.; et al. Changes in protein expression in two cholangiocarcinoma cell lines undergoing formation of multicellular tumor spheroids in vitro. PLoS ONE 2015. [Google Scholar] [CrossRef] [PubMed]
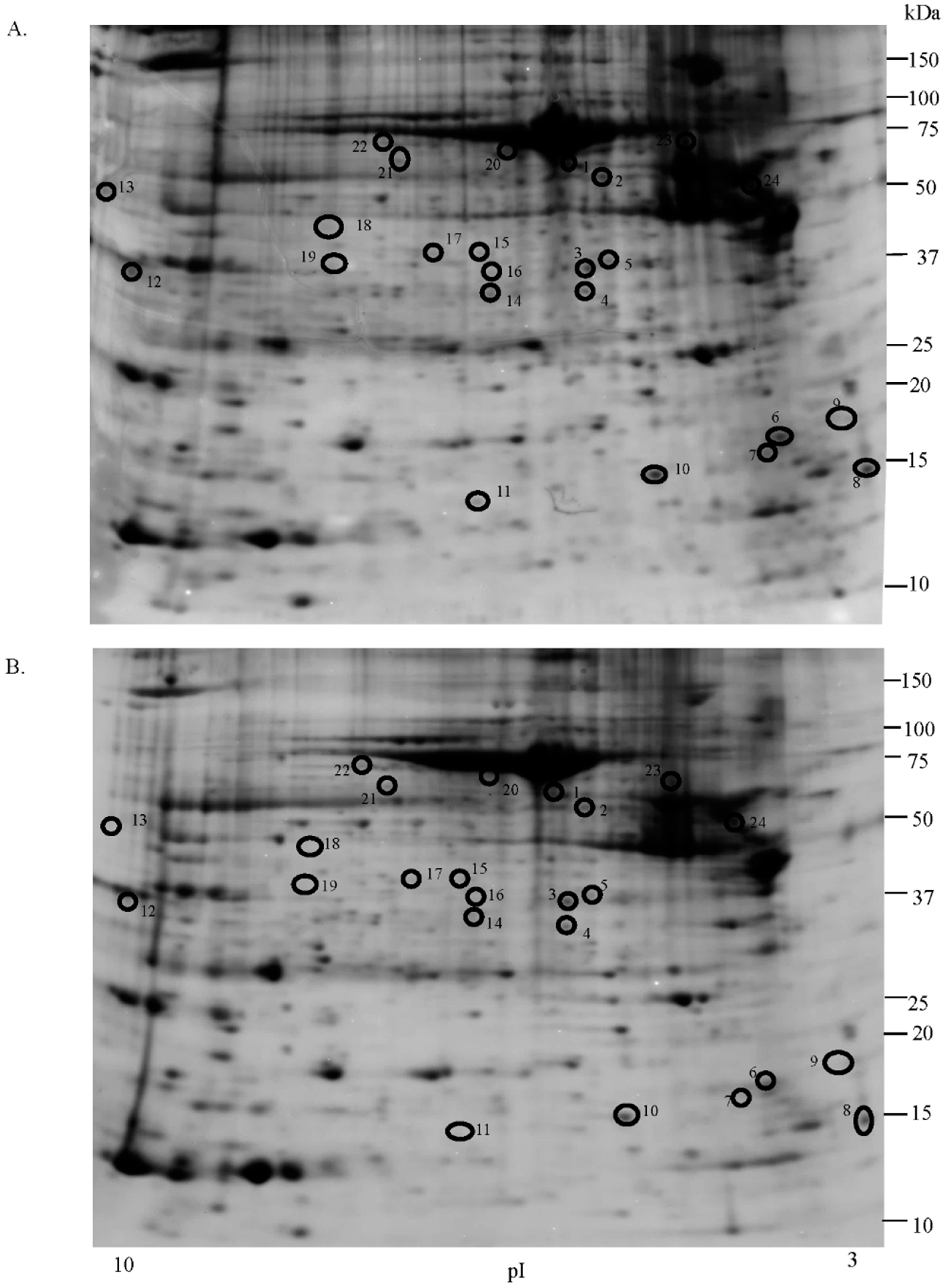
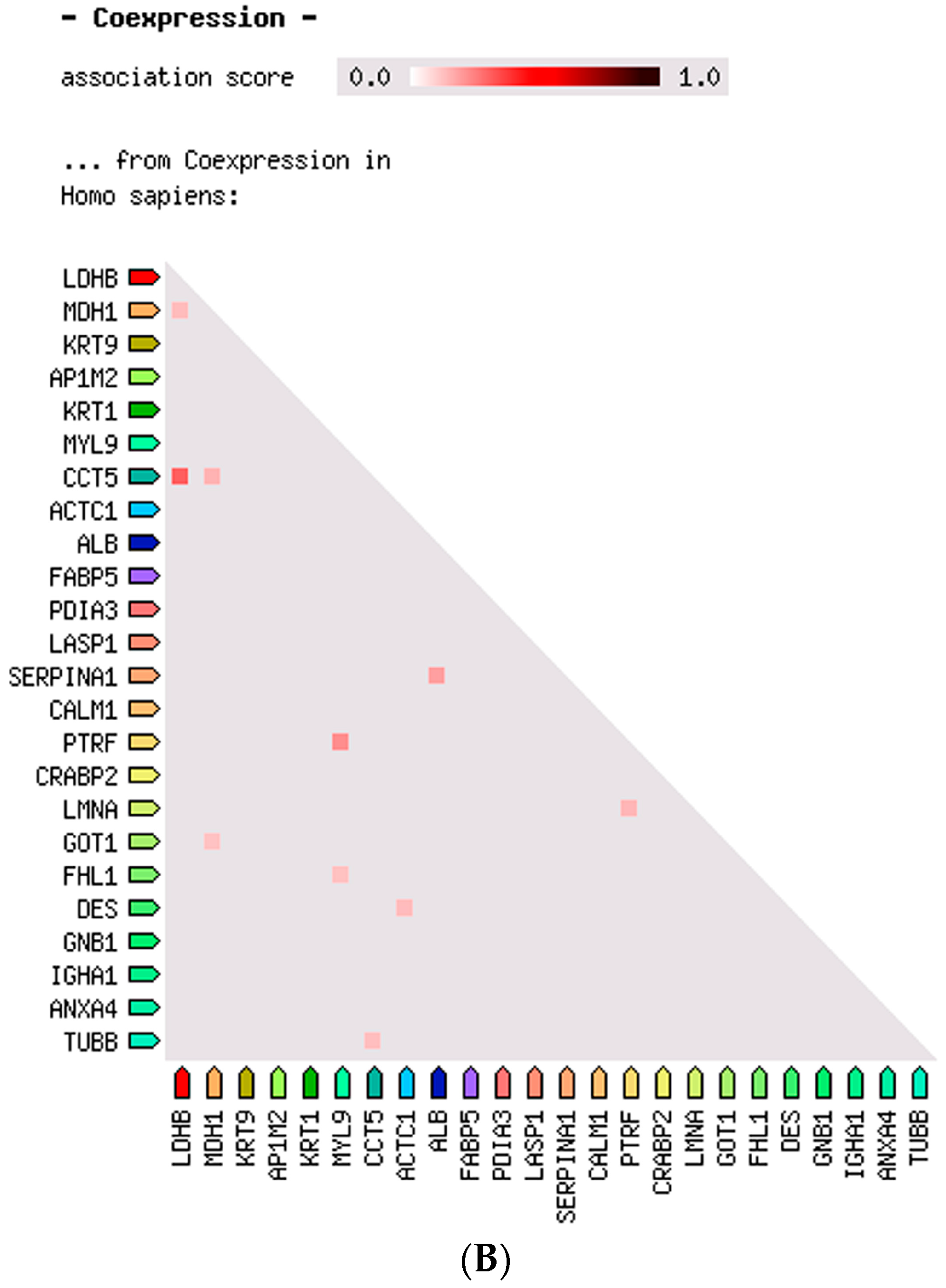
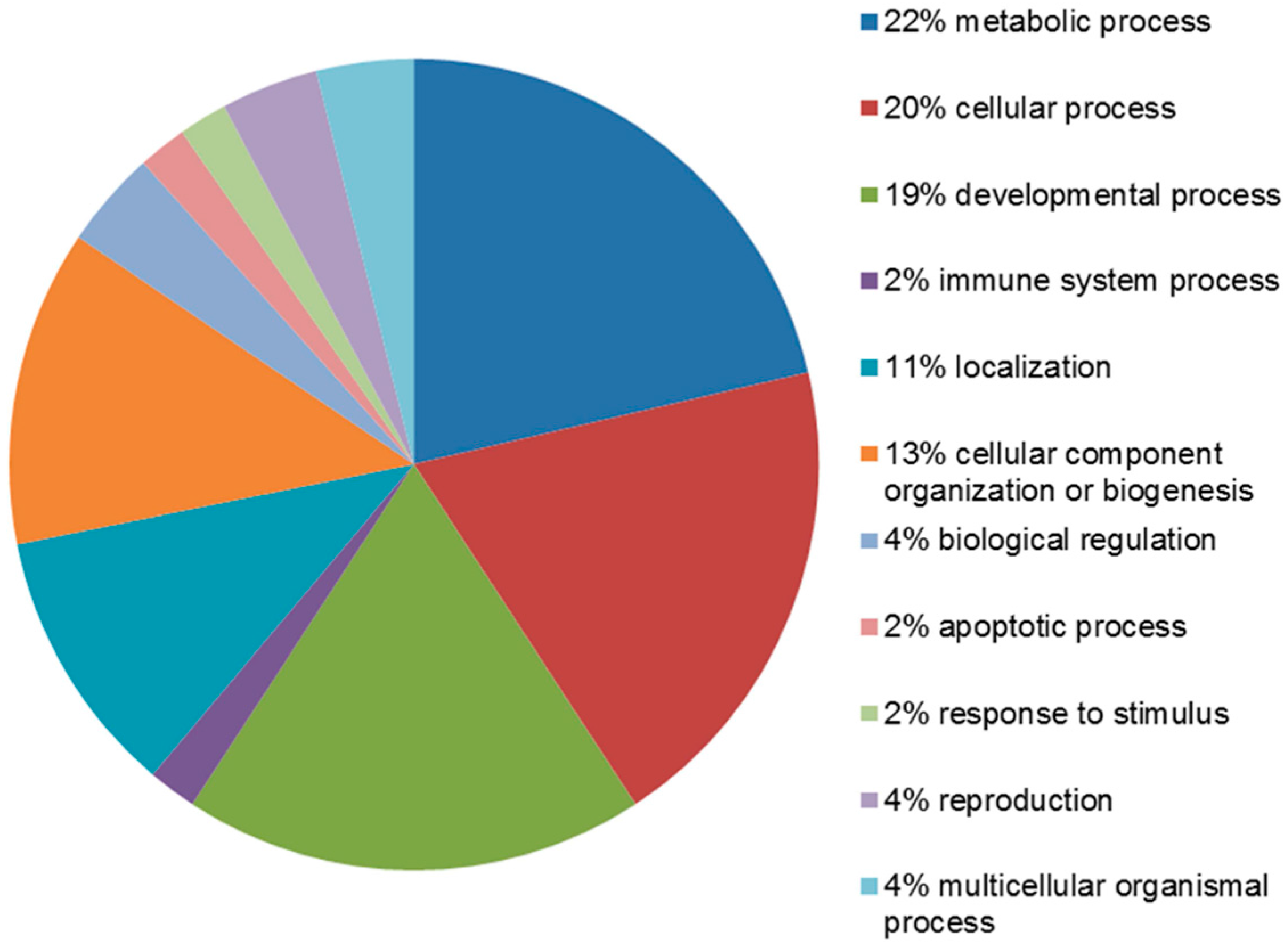
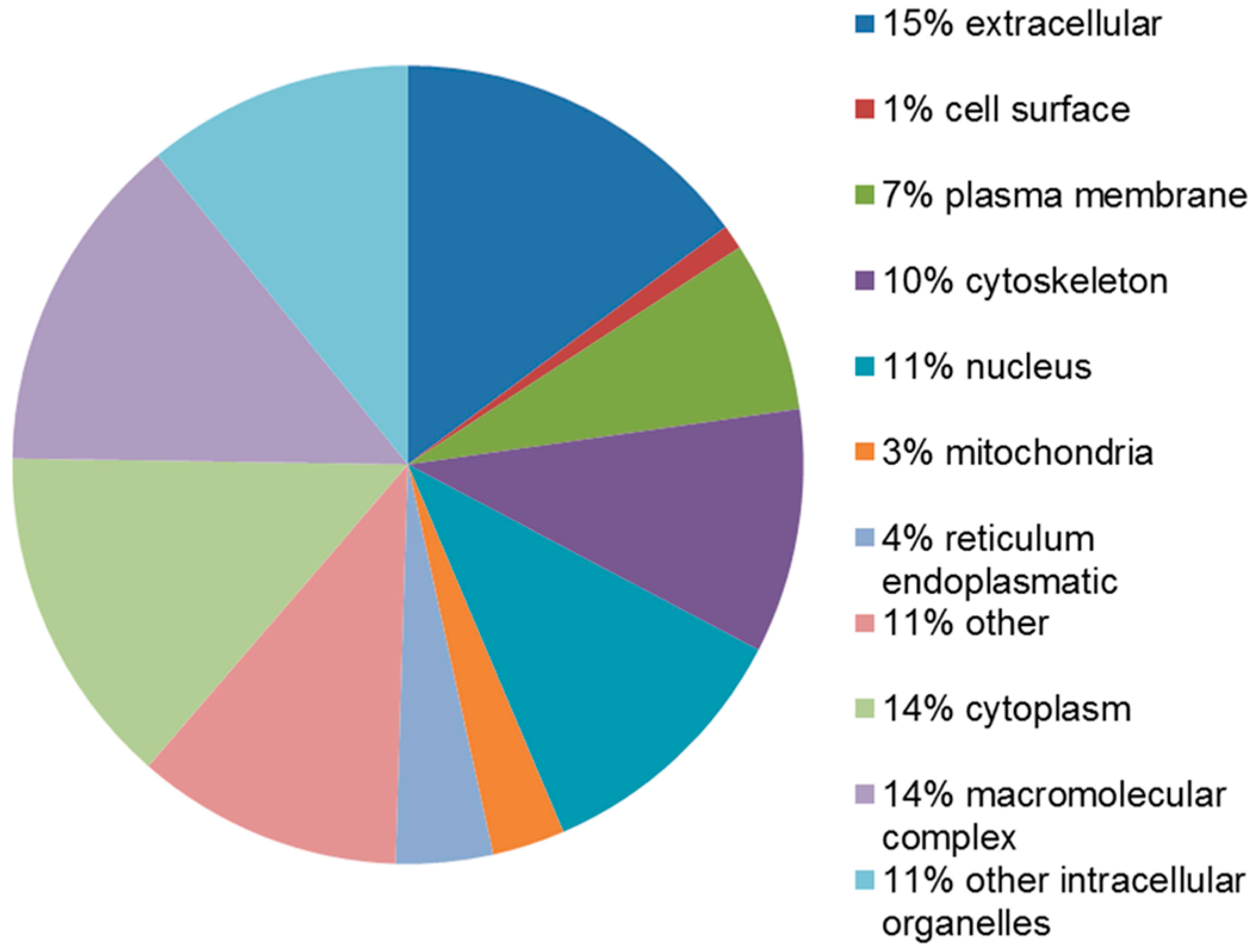
| Accession Number | Spot No. | Protein Description | Gene Symbol | Protein Score * | Fold Change ** | SEM *** | Subcellular Localization | p-Value | Adjusted p-Value **** |
|---|---|---|---|---|---|---|---|---|---|
| Metabolic process | |||||||||
| Lipid metabolic process | |||||||||
| Q01469 | 11 | Fatty acid binding protein, epidermal | FABP5 | 348 | 3.5 | 1.220 | Cytoplasm | 0.011 | 0.264 |
| P29373 | 10 | Cellular retinoic acid-binding protein 2 | CRABP5 | 145 | 2.25 | 0.397 | Cytoplasm | 0.011 | 0.264 |
| P09525 | 4 | Annexin A4 | ANXA4 | 2767 | 1.5 | 0.202 | Extracellular | 0.029 | 0.696 |
| Carbohydrate and TCA metabolic process | |||||||||
| P40925 | 19 | Malate dehydrogenase cytoplasmic | MDH1 | 45 | 2.4 | 0.691 | Cytoskeleton | 0.022 | 0.528 |
| P07195 | 3 | l-lactate dehydrogenase B chain | LDHB | 1526 | 1.5 | 0.241 | Cytoplasm | 0.014 | 0.336 |
| Amino acid metabolic process | |||||||||
| P17174 | 18 | Aspartate aminotransferase cytoplasmic | GOT1 | 225 | 4.3 | 0.444 | Cytoplasm | 0.018 | 0.432 |
| Nucleobase-containing compound metabolic process | |||||||||
| Q13642 | 12 | Four and a half LIM domains protein 1 | FHL1 | 224 | 5 | 2.338 | Cytoplasm | 0.022 | 0.528 |
| P62873 | 5 | isoform 2 of Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit β | GNB1 | 114 | 1.76 | 0.266 | Other cell body | 0.018 | 0.432 |
| Q6NZI2 | 9 | isoform 3 of polymerase I and transcript release factor | PTRF | 229 | 1.75 | 0.793 | Plasma membrane | 0.019 | 0.456 |
| Protein metabolic process | |||||||||
| P48643 | 17 | T-complex protein 1 subunit epsilon | CCT5 | 60 | 2.5 | 1.184 | Other cell body | 0.035 | 0.840 |
| P30101 | 1 | Protein disulfide isomerase A3 | PDIA3 | 161 | 1.6 | 0.359 | Cell surface | 0.019 | 0.456 |
| P09525 | 24 | α-1-antitrypsin | SERPINA1 | 335 | 1.5 | 0.832 | Endoplasmatic reticulum | 0.014 | 0.336 |
| Cellular process | |||||||||
| Cell communication, cycle and movement | |||||||||
| P68032 | 23 | Actin, α cardiac muscle 1 | ACTC1 | 248 | 5.1 | 1.527 | Cytoskeleton | 0.018 | 0.432 |
| P04264 | 13 | Keratin, type II cytoskeletal 1 | KRT1 | 107 | 3.1 | 1.821 | Extracellular | 0.022 | 0.528 |
| P02545 | 8 | Calmodulin 1 | CALM1 | 313 | 1.9 | 0.258 | Cytoskeleton | 0.011 | 0.264 |
| P07437 | 2 | Tubulin β chain | TUBB | 317 | 1.9 | 0.455 | Cytoskeleton | 0.011 | 0.264 |
| Developmental process | |||||||||
| Anatomical structure and morphogenesis | |||||||||
| P02545 | 22 | isoform 5 of prelamin-A/C | LMNA | 128 | 2.4 | 1.043 | Cytoplasm | 0.028 | 0.672 |
| P35527 | 14 | Keratin, type I cytoskeletal 9 | KRT9 | 214 | 1.9 | 0.288 | Extracellular | 0.035 | 0.840 |
| Cellular component organization or biogenesis | |||||||||
| Cellular component organization | |||||||||
| P24844 | 6 | Myosin regulatory light polypeptide 9 | MYL9 | 100 | 2 | 0.482 | Cytoplasm | 0.017 | 0.408 |
| P17661 | 7 | Desmin | DES | 190 | 1.7 | 0.626 | Cytoplasm | 0.035 | 0.840 |
| Transport | |||||||||
| P02768 | 21 | Serum albumin | ALB | 117 | 2.5 | 0.803 | Extracellular | 0.018 | 0.432 |
| Q9Y6Q5 | 16 | AP-1 complex subunit mu-2 | AP1M2 | 54 | 2.2 | 0.720 | Macromolecular complex | 0.045 | 1.080 |
| Immune system process | |||||||||
| P01876 | 20 | Immunoglobulin heavy constant α 1 | IGAH1 | 68 | 1.9 | 0.412 | Extracellular | 0.028 | 0.672 |
| Biological regulation | |||||||||
| Q14847 | 15 | LIM and SH3 domain protein 1 fragment | LASP1 | 66 | 1.7 | 0.444 | Cytoskeleton | 0.031 | 0.744 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ura, B.; Scrimin, F.; Arrigoni, G.; Franchin, C.; Monasta, L.; Ricci, G. A Proteomic Approach for the Identification of Up-Regulated Proteins Involved in the Metabolic Process of the Leiomyoma. Int. J. Mol. Sci. 2016, 17, 540. https://doi.org/10.3390/ijms17040540
Ura B, Scrimin F, Arrigoni G, Franchin C, Monasta L, Ricci G. A Proteomic Approach for the Identification of Up-Regulated Proteins Involved in the Metabolic Process of the Leiomyoma. International Journal of Molecular Sciences. 2016; 17(4):540. https://doi.org/10.3390/ijms17040540
Chicago/Turabian StyleUra, Blendi, Federica Scrimin, Giorgio Arrigoni, Cinzia Franchin, Lorenzo Monasta, and Giuseppe Ricci. 2016. "A Proteomic Approach for the Identification of Up-Regulated Proteins Involved in the Metabolic Process of the Leiomyoma" International Journal of Molecular Sciences 17, no. 4: 540. https://doi.org/10.3390/ijms17040540