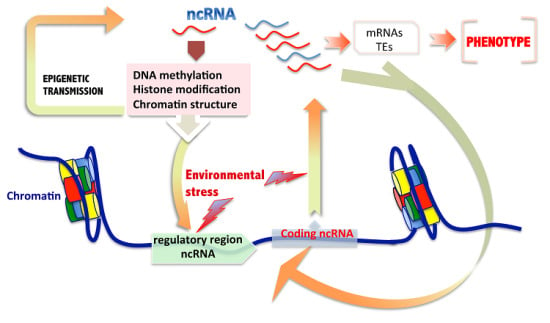
Role of Non-Coding RNAs in the Transgenerational Epigenetic Transmission of the Effects of Reprotoxicants
Abstract
:1. Introduction
2. Mechanisms of Epigenetic Transmission of Adverse Effects
2.1. DNA Methylation
2.2. Chromatin Histone Code
2.3. Non-Coding RNAs and Intermingled Epigenetic Mechanisms Involved in Transgenerational Transmission
3. Are ncRNAs Sufficient for Transgenerational Epigenetic Inheritance of Environmental Induced Characters?
4. RNA Epigenetics: A New Player Emerging on the Scene
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar]
- Consortium, F. A promoter-level mammalian expression atlas. Nature 2014, 507, 462–470. [Google Scholar] [Green Version]
- Pauli, A.; Rinn, J.L.; Schier, A.F. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 2011, 12, 136–149. [Google Scholar]
- Wilhelm, D.; Bernard, P. Non-Coding RNA and the Reproductive System; Springer Netherlands: Dordrecht, The Netherlands, 2016. [Google Scholar]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar]
- Harries, L.W. Long non-coding RNAs and human disease. Biochem. Soc. Trans. 2012, 40, 902–906. [Google Scholar]
- García-López, J.; Brieño-Enríquez, M.A.; del Mazo, J. MicroRNA biogenesis and variability. Biomol. Concepts 2013, 4, 367–380. [Google Scholar]
- Brown, C.J.; Hendrich, B.D.; Rupert, J.L.; Lafreniere, R.G.; Xing, Y.; Lawrence, J.; Willard, H.F. The human xist gene—Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 1992, 71, 527–542. [Google Scholar]
- Salido, E.C.; Yen, P.H.; Mohandas, T.K.; Shapiro, L.J. Expression of the X-inactivation-associated gene XIST during spermatogenesis. Nat. Genet. 1992, 2, 196–199. [Google Scholar]
- McCarrey, J.R.; Dilworth, D.D. Expression of XIST in mouse germ cells correlates with X-chromosome inactivation. Nat. Genet. 1992, 2, 200–203. [Google Scholar]
- Lee, J.T. Lessons from X-chromosome inactivation: Long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009, 23, 1831–1842. [Google Scholar]
- Koerner, M.V.; Pauler, F.M.; Huang, R.; Barlow, D.P. The function of non-coding RNAs in genomic imprinting. Development 2009, 136, 1771–1783. [Google Scholar]
- Berger, S.L.; Kouzarides, T.; Shiekhattar, R.; Shilatifard, A. An operational definition of epigenetics. Genes Dev. 2009, 23, 781–783. [Google Scholar]
- Feil, R.; Fraga, M.F. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2011, 13, 97–109. [Google Scholar]
- Skakkebaek, N.E.; Rajpert-De Meyts, E.; Main, K.M. Testicular dysgenesis syndrome: An increasingly common developmental disorder with environmental aspects. Hum. Reprod. 2001, 16, 972–978. [Google Scholar]
- Asklund, C.; Jorgensen, N.; Kold Jensen, T.; Skakkebaek, N.E. Biology and epidemiology of testicular dysgenesis syndrome. BJU Int. 2004, 93, 6–11. [Google Scholar]
- Hou, L.; Zhang, X.; Wang, D.; Baccarelli, A. Environmental chemical exposures and human epigenetics. Int. J. Epidemiol. 2012, 41, 79–105. [Google Scholar]
- WHO-UNEP. State of the Science of Endocrine Disrupting Chemicals. In State of the Science of Endocrine Disrupting Chemicals; Bergman, A., Heindel, J.J., Jobling, S., Kidd, K.A., Zoeller, R.T., Eds.; World Health Organization: Geneva, Switzerland, 2012. [Google Scholar]
- Lamb, J.C.T.; Boffetta, P.; Foster, W.G.; Goodman, J.E.; Hentz, K.L.; Rhomberg, L.R.; Staveley, J.; Swaen, G.; van Der Kraak, G.; Williams, A.L. Comments on the opinions published by Bergman et al. (2015) on Critical comments on the WHO-UNEP state of the science of endocrine disrupting chemicals (Lamb et al. 2014). Regul. Toxicol. Pharmacol. 2015, 73, 754–757. [Google Scholar]
- Bergman, A.; Becher, G.; Blumberg, B.; Bjerregaard, P.; Bornman, R.; Brandt, I.; Casey, S.C.; Frouin, H.; Giudice, L.C.; Heindel, J.J.; et al. Manufacturing doubt about endocrine disrupter science—A rebuttal of industry-sponsored critical comments on the UNEP/WHO report “State of the Science of Endocrine Disrupting Chemicals 2012”. Regul. Toxicol. Pharmacol. 2015, 73, 1007–1017. [Google Scholar]
- Del Mazo, J.; Brieno-Enriquez, M.A.; Garcia-Lopez, J.; Lopez-Fernandez, L.A.; de Felici, M. Endocrine disruptors, gene deregulation and male germ cell tumors. Int. J. Dev. Biol. 2013, 57, 225–239. [Google Scholar] [PubMed]
- Nilsson, E.E.; Skinner, M.K. Environmentally induced epigenetic transgenerational inheritance of reproductive disease. Biol. Reprod. 2015, 93, 145. [Google Scholar] [PubMed]
- Heard, E.; Martienssen, R.A. Transgenerational epigenetic inheritance: Myths and mechanisms. Cell 2014, 157, 95–109. [Google Scholar] [PubMed]
- Skinner, M.K. Endocrine disruptors in 2015: Epigenetic transgenerational inheritance. Nat. Rev. Endocrinol. 2016, 12, 68–70. [Google Scholar] [PubMed]
- Lopez-Casas, P.P.; Mizrak, S.C.; Lopez-Fernandez, L.A.; Paz, M.; de Rooij, D.G.; del Mazo, J. The effects of different endocrine disruptors defining compound-specific alterations of gene expression profiles in the developing testis. Reprod. Toxicol. 2012, 33, 106–115. [Google Scholar] [Green Version]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [PubMed]
- Wu, S.C.; Zhang, Y. Active DNA demethylation: Many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010, 11, 607–620. [Google Scholar] [PubMed]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [PubMed]
- Guibert, S.; Forne, T.; Weber, M. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res. 2012, 22, 633–641. [Google Scholar] [PubMed]
- Seisenberger, S.; Andrews, S.; Krueger, F.; Arand, J.; Walter, J.; Santos, F.; Popp, C.; Thienpont, B.; Dean, W.; Reik, W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 2012, 48, 849–862. [Google Scholar] [PubMed]
- Kafri, T.; Ariel, M.; Brandeis, M.; Shemer, R.; Urven, L.; Mc Carrey, J.; Cedar, H.; Razin, A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992, 6, 705–714. [Google Scholar] [PubMed]
- Del Mazo, J.; Prantera, G.; Torres, M.; Ferraro, M. DNA methylation changes during mouse spermatogenesis. Chromosome Res. 1994, 2, 147–152. [Google Scholar] [PubMed]
- Feil, R. Epigenetic asymmetry in the zygote and mammalian development. Int. J. Dev. Biol. 2009, 53, 191–201. [Google Scholar] [PubMed]
- Lane, N.; Dean, W.; Erhardt, S.; Hajkova, P.; Surani, A.; Walter, J.; Reik, W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 2003, 35, 88–93. [Google Scholar] [PubMed]
- Smith, Z.D.; Chan, M.M.; Humm, K.C.; Karnik, R.; Mekhoubad, S.; Regev, A.; Eggan, K.; Meissner, A. DNA methylation dynamics of the human preimplantation embryo. Nature 2014, 511, 611–615. [Google Scholar] [PubMed]
- Herbst, A.L.; Ulfelder, H.; Poskanzer, D.C. Adenocarcinoma of the vagina. Association of maternal stilbestrol therapy with tumor appearance in young women. N. Engl. J. Med. 1971, 284, 878–881. [Google Scholar] [PubMed]
- Newbold, R.R. Lessons learned from perinatal exposure to diethylstilbestrol. Toxicol. Appl. Pharmacol. 2004, 199, 142–150. [Google Scholar] [PubMed]
- Sato, K.; Fukata, H.; Kogo, Y.; Ohgane, J.; Shiota, K.; Mori, C. Neonatal exposure to diethylstilbestrol alters expression of DNA methyltransferases and methylation of genomic DNA in the mouse uterus. Endocr. J. 2009, 56, 131–139. [Google Scholar] [PubMed]
- Edwards, T.M.; Myers, J.P. Environmental exposures and gene regulation in disease etiology. Environ. Health Perspect. 2007, 115, 1264–1270. [Google Scholar] [PubMed]
- del Mazo, J.; Garcia-Lopez, J.; Weber, M. Epigenetic traits of testicular cancer: From primordial germ cells to germ cell tumors. Epigenomics 2014, 6, 253–255. [Google Scholar] [PubMed]
- Manikkam, M.; Tracey, R.; Guerrero-Bosagna, C.; Skinner, M.K. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS ONE 2013, 8, e55387. [Google Scholar]
- Manikkam, M.; Guerrero-Bosagna, C.; Tracey, R.; Haque, M.M.; Skinner, M.K. Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures. PLoS ONE 2012, 7, e31901. [Google Scholar]
- Manikkam, M.; Haque, M.M.; Guerrero-Bosagna, C.; Nilsson, E.E.; Skinner, M.K. Pesticide methoxychlor promotes the epigenetic transgenerational inheritance of adult-onset disease through the female germline. PLoS ONE 2014, 9, e102091. [Google Scholar]
- Osada, S.; Nishikawa, J.; Nakanishi, T.; Tanaka, K.; Nishihara, T. Some organotin compounds enhance histone acetyltransferase activity. Toxicol. Lett. 2005, 155, 329–335. [Google Scholar]
- Tabb, M.M.; Blumberg, B. New modes of action for endocrine-disrupting chemicals. Mol. Endocrinol. 2006, 20, 475–482. [Google Scholar]
- Hammoud, S.S.; Nix, D.A.; Zhang, H.; Purwar, J.; Carrell, D.T.; Cairns, B.R. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009, 460, 473–478. [Google Scholar]
- Seki, Y.; Hayashi, K.; Itoh, K.; Mizugaki, M.; Saitou, M.; Matsui, Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev. Biol. 2005, 278, 440–458. [Google Scholar]
- Lees-Murdock, D.J.; De Felici, M.; Walsh, C.P. Methylation dynamics of repetitive DNA elements in the mouse germ cell lineage. Genomics 2003, 82, 230–237. [Google Scholar]
- Stewart, K.R.; Veselovska, L.; Kim, J.; Huang, J.; Saadeh, H.; Tomizawa, S.; Smallwood, S.A.; Chen, T.; Kelsey, G. Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes. Dev. 2015, 29, 2449–2462. [Google Scholar]
- Yan, W. Potential roles of noncoding RNAs in environmental epigenetic transgenerational inheritance. Mol. Cell. Endocrinol. 2014, 398, 24–30. [Google Scholar]
- Holoch, D.; Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar]
- Guerin, T.M.; Palladino, F.; Robert, V.J. Transgenerational functions of small RNA pathways in controlling gene expression in C. elegans. Epigenetics 2014, 9, 37–44. [Google Scholar]
- Ashe, A.; Sapetschnig, A.; Weick, E.M.; Mitchell, J.; Bagijn, M.P.; Cording, A.C.; Doebley, A.L.; Goldstein, L.D.; Lehrbach, N.J.; le Pen, J.; et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 2012, 150, 88–99. [Google Scholar]
- Rajasethupathy, P.; Antonov, I.; Sheridan, R.; Frey, S.; Sander, C.; Tuschl, T.; Kandel, E.R. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 2012, 149, 693–707. [Google Scholar]
- Miousse, I.R.; Chalbot, M.C.; Lumen, A.; Ferguson, A.; Kavouras, I.G.; Koturbash, I. Response of transposable elements to environmental stressors. Mutat. Res. Rev. Mutat. Res. 2015, 765, 19–39. [Google Scholar]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar]
- Garcia-Lopez, J.; Hourcade Jde, D.; Alonso, L.; Cardenas, D.B.; del Mazo, J. Global characterization and target identification of piRNAs and endo-siRNAs in mouse gametes and zygotes. Biochim. Biophys. Acta 2014, 1839, 463–475. [Google Scholar]
- Garcia-Lopez, J.; Alonso, L.; Cardenas, D.B.; Artaza-Alvarez, H.; Hourcade Jde, D.; Martinez, S.; Brieno-Enriquez, M.A.; del Mazo, J. Diversity and functional convergence of small noncoding RNAs in male germ cell differentiation and fertilization. RNA 2015, 21, 946–962. [Google Scholar]
- Suzuki, S.; Shaw, G.; Kaneko-Ishino, T.; Ishino, F.; Renfree, M.B. The evolution of mammalian genomic imprinting was accompanied by the acquisition of novel CpG islands. Genome Biol. Evol. 2011, 3, 1276–1283. [Google Scholar]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013, 14, 699–712. [Google Scholar]
- Quinodoz, S.; Guttman, M. Long noncoding RNAs: An emerging link between gene regulation and nuclear organization. Trends Cell Biol. 2014, 24, 651–663. [Google Scholar]
- Bonasio, R.; Shiekhattar, R. Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 2014, 48, 433–455. [Google Scholar]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar]
- Kornienko, A.E.; Dotter, C.P.; Guenzl, P.M.; Gisslinger, H.; Gisslinger, B.; Cleary, C.; Kralovics, R.; Pauler, F.M.; Barlow, D.P. Long non-coding RNAs display higher natural expression variation than protein-coding genes in healthy humans. Genome Biol. 2016, 17, 14. [Google Scholar]
- Amin, V.; Harris, R.A.; Onuchic, V.; Jackson, A.R.; Charnecki, T.; Paithankar, S.; Lakshmi Subramanian, S.; Riehle, K.; Coarfa, C.; Milosavljevic, A. Epigenomic footprints across 111 reference epigenomes reveal tissue-specific epigenetic regulation of lincRNAs. Nat. Commun. 2015, 6, 6370. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Dhir, A.; Dhir, S.; Proudfoot, N.J.; Jopling, C.L. Microprocessor mediates transcriptional termination of long noncoding RNA transcripts hosting microRNAs. Nat. Struct. Mol. Biol. 2015, 22, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.; Gaidatzis, D.; Pfeffer, S.; Lagos-Quintana, M.; Landgraf, P.; Iovino, N.; Morris, P.; Brownstein, M.J.; Kuramochi-Miyagawa, S.; Nakano, T.; et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006, 442, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Hirakata, S.; Siomi, M.C. piRNA biogenesis in the germline: From transcription of piRNA genomic sources to piRNA maturation. Biochim. Biophys. Acta 2016, 1859, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Olovnikov, I.A.; Kalmykova, A.I. piRNA clusters as a main source of small RNAs in the animal germline. Biochemistry 2013, 78, 572–584. [Google Scholar] [CrossRef] [PubMed]
- Kapusta, A.; Kronenberg, Z.; Lynch, V.J.; Zhuo, X.; Ramsay, L.; Bourque, G.; Yandell, M.; Feschotte, C. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 2013, 9, e1003470. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Cheng, E.C.; Zhong, M.; Lin, H. Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res. 2015, 25, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Ha, H.; Song, J.; Wang, S.; Kapusta, A.; Feschotte, C.; Chen, K.C.; Xing, J. A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genom. 2014, 15, 545. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Chen, X.; Zhang, X.; Duan, X.; Pan, T.; Hu, Q.; Zhang, Y.; Zhong, F.; Liu, J.; Zhang, H.; et al. An Lnc RNA (GAS5)/SnoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes. Nucleic Acids Res. 2015, 43, 3712–3725. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Zhao, J.C.; Kim, J.; Fong, K.W.; Yang, Y.A.; Chakravarti, D.; Mo, Y.Y.; Yu, J. LncRNA HOTAIR Enhances the Androgen-Receptor-Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer. Cell Rep. 2015, 13, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Gibb, E.A.; Brown, C.J.; Lam, W.L. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer 2011, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Bhan, A.; Hussain, I.; Ansari, K.I.; Bobzean, S.A.; Perrotti, L.I.; Mandal, S.S. Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J. Steroid Biochem. 2014, 141, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Reczko, M.; Maragkakis, M.; Dalamagas, T.M.; Hatzigeorgiou, A.G. DIANA-LncBase: Experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 2013, 41, D239–D245. [Google Scholar] [CrossRef] [PubMed]
- Paci, P.; Colombo, T.; Farina, L. Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst. Biol. 2014, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Chiyomaru, T.; Yamamura, S.; Fukuhara, S.; Yoshino, H.; Kinoshita, T.; Majid, S.; Saini, S.; Chang, I.; Tanaka, Y.; Enokida, H.; et al. Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PLoS ONE 2013, 8, e70372. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xue, Y. Emerging roles of non-coding RNAs in epigenetic regulation. Sci. China Life Sci 2016, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Anway, M.D.; Cupp, A.S.; Uzumcu, M.; Skinner, M.K. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 2005, 308, 1466–1469. [Google Scholar] [CrossRef] [PubMed]
- Stouder, C.; Paoloni-Giacobino, A. Transgenerational effects of the endocrine disruptor vinclozolin on the methylation pattern of imprinted genes in the mouse sperm. Reproduction 2010, 139, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Bosagna, C.; Covert, T.R.; Haque, M.M.; Settles, M.; Nilsson, E.E.; Anway, M.D.; Skinner, M.K. Epigenetic transgenerational inheritance of vinclozolin induced mouse adult onset disease and associated sperm epigenome biomarkers. Reprod. Toxicol. 2012, 34, 694–707. [Google Scholar] [CrossRef] [PubMed]
- Brieno-Enriquez, M.A.; Garcia-Lopez, J.; Cardenas, D.B.; Guibert, S.; Cleroux, E.; Ded, L.; Hourcade Jde, D.; Peknicova, J.; Weber, M.; del Mazo, J. Exposure to endocrine disruptor induces transgenerational epigenetic deregulation of microRNAs in primordial germ cells. PLoS ONE 2015, 10, e0124296. [Google Scholar]
- Chen, Q.; Yan, M.; Cao, Z.; Li, X.; Zhang, Y.; Shi, J.; Feng, G.H.; Peng, H.; Zhang, X.; Zhang, Y.; et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016, 351, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Silva, M.R.; Frugier, M.; Tosar, J.P.; Correa-Dominguez, A.; Ronalte-Alves, L.; Parodi-Talice, A.; Rovira, C.; Robello, C.; Goldenberg, S.; Cayota, A. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol. Biochem. Parasitol. 2010, 171, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Shigematsu, M.; Kirino, Y. tRNA-derived short non-coding RNA as interacting partners of argonaute proteins. Gene Regul. Syst. Biol. 2015, 9, 27–33. [Google Scholar]
- Keam, S.P.; Young, P.E.; McCorkindale, A.L.; Dang, T.H.; Clancy, J.L.; Humphreys, D.T.; Preiss, T.; Hutvagner, G.; Martin, D.I.; Cropley, J.E.; et al. The human Piwi protein HIWI2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res. 2014, 42, 8984–8995. [Google Scholar] [CrossRef] [PubMed]
- Heyns, M.; Kovalchuk, O. Non-coding RNAs including miRNAs, piRNAs, and tRNAs in human cancer. Oncotarget 2015, 6, 23055–23057. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014, 15, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S.; et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 2015, 347, 1002–1006. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Hao, Y.J.; Zhang, Y.; Li, M.M.; Wang, M.; Han, W.; Wu, Y.; Lv, Y.; Hao, J.; Wang, L.; et al. m6A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 2015, 16, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Youngren, K.K.; Coveney, D.; Peng, X.; Bhattacharya, C.; Schmidt, L.S.; Nickerson, M.L.; Lamb, B.T.; Deng, J.M.; Behringer, R.R.; Capel, B.; et al. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature 2005, 435, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002, 71, 817–846. [Google Scholar] [CrossRef] [PubMed]
- Keegan, L.P.; Leroy, A.; Sproul, D.; O’Connell, M.A. Adenosine deaminases acting on RNA (ADARs): RNA-editing enzymes. Genome Biol. 2004, 5, 209. [Google Scholar] [CrossRef] [PubMed]
- Hogg, M.; Paro, S.; Keegan, L.P.; O’Connell, M.A. RNA editing by mammalian ADARs. Adv. Genet. 2011, 73, 87–120. [Google Scholar] [PubMed]
- Wahlstedt, H.; Daniel, C.; Enstero, M.; Ohman, M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009, 19, 978–986. [Google Scholar] [CrossRef] [PubMed]
- Alon, S.; Mor, E.; Vigneault, F.; Church, G.M.; Locatelli, F.; Galeano, F.; Gallo, A.; Shomron, N.; Eisenberg, E. Systematic identification of edited microRNAs in the human brain. Genome Res. 2012, 22, 1533–1540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Lopez, J.; Hourcade Jde, D.; del Mazo, J. Reprogramming of microRNAs by adenosine-to-inosine editing and the selective elimination of edited microRNA precursors in mouse oocytes and preimplantation embryos. Nucleic Acids Res. 2013, 41, 5483–5493. [Google Scholar] [CrossRef] [PubMed]
- Gommans, W.M. A-to-I editing of microRNAs: Regulating the regulators? Semin. Cell Dev. Biol. 2012, 23, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Stocker, J.; Huang, H.W.; Wang, H.M.; Chang, H.W.; Chiu, C.C.; Cho, C.L.; Tseng, C.N. Reduction of RNA A-to-I editing in Drosophila acclimated to heat shock. J. Med. Sci. 2013, 29, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Khermesh, K.; D’Erchia, A.M.; Barak, M.; Annese, A.; Wachtel, C.; Levanon, E.Y.; Picardi, E.; Eisenberg, E. Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer’s disease. RNA 2015, 22, 290–302. [Google Scholar] [CrossRef] [PubMed]
- El Yacoubi, B.; Bailly, M.; de Crecy-Lagard, V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012, 46, 69–95. [Google Scholar] [CrossRef] [PubMed]
- McCarrey, J.R. Distinctions between transgenerational and non-transgenerational epimutations. Mol. Cell. Endocrinol. 2014, 398, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K.; Guerrero-Bosagna, C.; Haque, M.M. Environmentally induced epigenetic transgenerational inheritance of sperm epimutations promote genetic mutations. Epigenetics 2015, 10, 762–771. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larriba, E.; Del Mazo, J. Role of Non-Coding RNAs in the Transgenerational Epigenetic Transmission of the Effects of Reprotoxicants. Int. J. Mol. Sci. 2016, 17, 452. https://doi.org/10.3390/ijms17040452
Larriba E, Del Mazo J. Role of Non-Coding RNAs in the Transgenerational Epigenetic Transmission of the Effects of Reprotoxicants. International Journal of Molecular Sciences. 2016; 17(4):452. https://doi.org/10.3390/ijms17040452
Chicago/Turabian StyleLarriba, Eduardo, and Jesús Del Mazo. 2016. "Role of Non-Coding RNAs in the Transgenerational Epigenetic Transmission of the Effects of Reprotoxicants" International Journal of Molecular Sciences 17, no. 4: 452. https://doi.org/10.3390/ijms17040452