Development and Characterization of Novel Microsatellite Markers for the Peach Fruit Moth Carposina sasakii (Lepidoptera: Carposinidae) Using Next-Generation Sequencing
Abstract
:1. Introduction
2. Results and Discussion
2.1. Microsatellite Marker Development
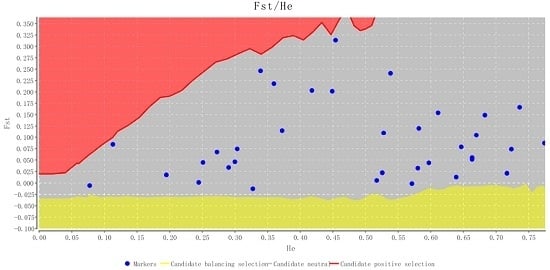
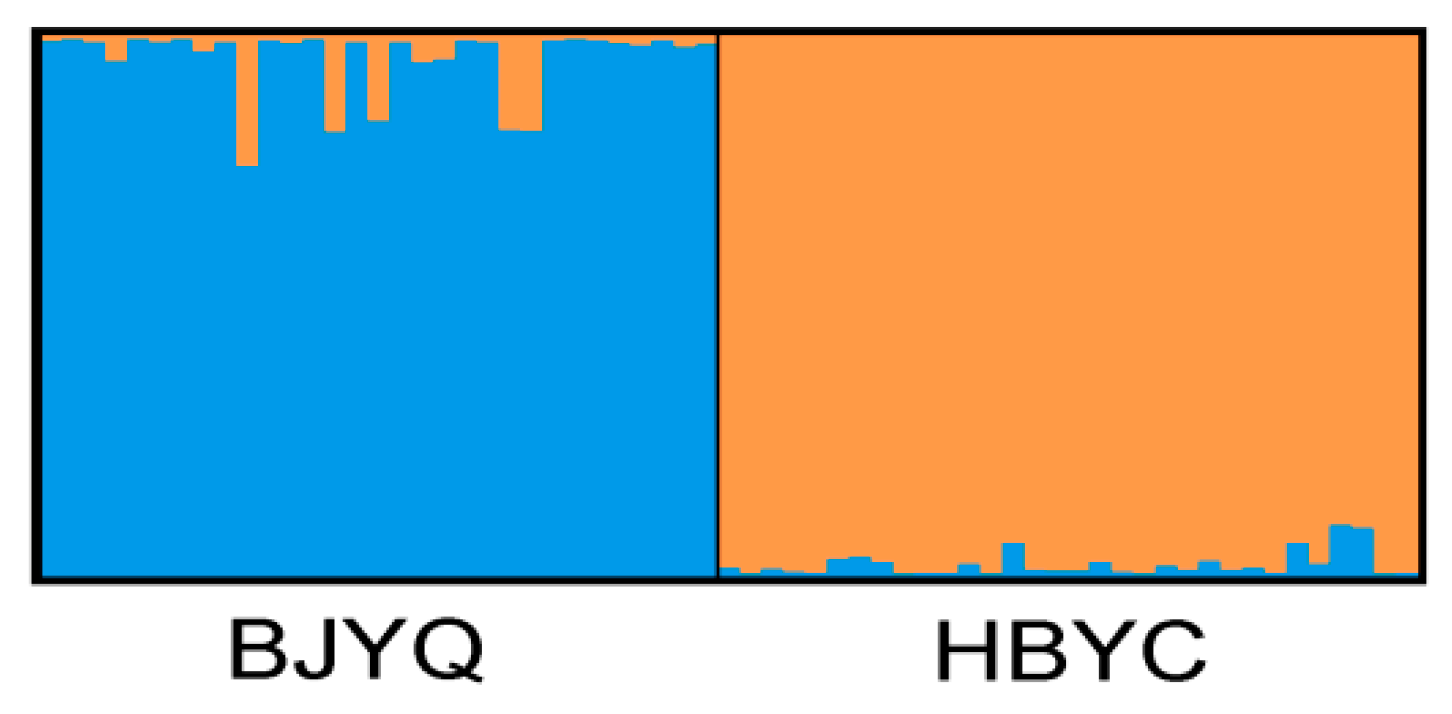
2.2. Characteristics of Validated Microsatellite Loci
3. Materials and Methods
3.1. Sample Collection and DNA Extraction
3.2. Sequencing, Microsatellites Searching and Primer Design
3.3. Primer Testing and Polymorphism Detection
3.4. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kim, D.S.; Lee, J.H.; Yiem, M.S. Spring emergence pattern of Carposina sasakii (Lepidoptera: Carposinidae) in apple orchards in Korea and its forecasting models based on degree-days. Environ. Entomol. 2000, 29, 1188–1198. [Google Scholar] [CrossRef]
- Liu, Y.S.; Cheng, J.A.; Mou, J.Y. Review of the advances of the peach fruit-borer (Carposina sasakii Matsmura). J. Shandong Agric. Univ. 1997, 28, 113–120. [Google Scholar]
- Han, X.J. The damage and prevention of peach friut moth on pomegranate. China Rurac Sci. Technol. 2002, 5, 17. [Google Scholar]
- Hua, B.Z.; Zhang, A.J.; Lu, X.Z.; Hua, L. Seasonal history of Carposina sasakii Matsumura on apricot orchard in Qinling mountain region, Shaanxi. Acta Agric. Boreali-Occident. Sin. 1998, 7, 32–35. [Google Scholar]
- Jiao, R.L. The occurrence and prevention of peach fruit moth. Plant Dr. 2006, 19, 19. [Google Scholar]
- Wang, X.M.; Liu, M. A detailed study of the living history, medicament prevention, cure and its relations with meteorology. J. Shenyang Univ. 1999, 63–71. (In Chinese) [Google Scholar]
- Cai, P.; Ding, W.Z. Briefing of peach fruit moth damage on pomegranate. China Fruits 1990, 35–37. (In Chinese) [Google Scholar]
- Hua, L.; Shen, B.C. Preliminary study on the peach fruit borer of wild jujube. Plant Prot. Sci. 1992, 15–17. (In Chinese) [Google Scholar]
- Hua, L.; Hua, B.Z.; Huang, W.L. The bionomics of peach fruit borer damaged on apricot tree. Acta Phytophylacica Sin. 1998, 25, 141–144. [Google Scholar]
- Hua, L.; Hua, B.Z. Preliminary study on the host-biotypes of peach fruit borer. Acta Phytophylacica Sin. 1995, 22, 165–170. [Google Scholar]
- Hua, L.; Wang, J.F.; Hui, X.X. The esterase isozymes of the Carposina sasakii from different hosts. Acta Agric. Boreali-Occident. Sin. 1995, 4, 95–96. [Google Scholar]
- Xu, Q.G.; Hua, B.Z. RAPD analysis on the speciation in host races of Carposina sasakii Matsumura (Lepidoptera: Carposinidae). Acta Entomol. Sin. 2004, 47, 379–383. [Google Scholar]
- Wang, J.; Yu, Y.; Li, L.L.; Guo, D.; Tao, Y.L.; Chu, D. Carposina sasakii (Lepidoptera: Carposinidae) in its native range consists of two sympatric cryptic lineages as revealed by mitochondrial COI gene sequences. J. Insect Sci. 2015, 15, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, G.; Nagai, S.; Lian, C.L.; Yamaguchi, H.; Sakamoto, S.; Yoshimatsu, S.; Oyama, K.; Itakura, S.; Yamaguchi, M. Development of compound microsatellite markers in the harmful red tide species Chattonella ovata (Raphidophyceae). Mol. Ecol. Notes 2007, 7, 1251–1253. [Google Scholar] [CrossRef]
- Bruford, M.W. Microsatellite and their application to population genetic studies. Curr. Opin. Genet. Dev. 1993, 3, 939–943. [Google Scholar] [CrossRef]
- Wei, S.J.; Cao, L.J.; Gong, Y.J.; Shi, B.C.; Wang, S.; Zhang, F.; Guo, X.J.; Wang, Y.M.; Chen, X.X. Population genetic structure and approximate Bayesian computation analyses reveal the southern origin and northward dispersal of the oriental fruit moth Grapholita molesta (Lepidoptera: Tortricidae) in its native range. Mol. Ecol. 2015, 24, 4094–4111. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.J.; Shi, B.C.; Gong, Y.J.; Jin, G.H.; Chen, X.X.; Meng, X.F. Genetic structure and demographic history reveal migration of the diamondback moth Plutella xylostella (Lepidoptera: Plutellidae) from the southern to northern regions of China. PLoS ONE 2013, 8, e59654. [Google Scholar] [CrossRef] [PubMed]
- Zalapa, J.E.; Cuevas, H.; Zhu, H.; Steffan, S.; Senalik, D.; Zeldin, E.; McCown, B.; Harbut, R.; Simon, P. Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences. Am. J. Bot. 2012, 99, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Fang, L.; Zhang, X.; Hu, J.; Bao, S.; Hao, J.; Li, L.; He, Y.; Jiang, J.; Wang, F.; Tian, S.; Zong, X. High-throughput development of SSR markers from pea (Pisum sativum L.) based on next generation sequencing of a purified chinese commercial variety. PLoS ONE 2015, 10, e0139775. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.M.; Sun, J.T.; Xue, X.F.; Zhu, W.C.; Hong, X.Y. Development and characterization of 18 novel EST-SSRs from the western flower thrips, Frankliniella occidentalis (Pergande). Int. J. Mol. Sci. 2012, 13, 2863–2876. [Google Scholar] [CrossRef] [PubMed]
- Malausa, T.; Gilles, A.; MeglÉCz, E.; Blanquart, H.; Duthoy, S.; Costedoat, C.; Dubut, V.; Pech, N.; Castagnone-Sereno, P.; DÉLye, C.; et al. High-throughput microsatellite isolation through 454 GS-FLX Titanium pyrosequencing of enriched DNA libraries. Mol. Ecol. Resour. 2011, 11, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010, 11, 485. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience 2012, 1, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.H.; Han, S.H.; Park, S.J. Development of seven microsatellite markers using next generation sequencing for the conservation on the Korean population of Dorcus hopei (E. Saunders, 1854) (Coleoptera, Lucanidae). Int. J. Mol. Sci. 2015, 16, 21330–21341. [Google Scholar] [CrossRef] [PubMed]
- Berman, M.; Austin, C.M.; Miller, A.D. Characterisation of the complete mitochondrial genome and 13 microsatellite loci through next-generation sequencing for the New Caledonian spider-ant Leptomyrmex pallens. Mol. Biol. Rep. 2014, 41, 1179–1187. [Google Scholar] [CrossRef] [PubMed]
- Nam, H.Y.; Coates, B.; Kim, K.S.; Park, M.; Lee, J.-H. Characterization of 12 novel microsatellite markers of Sogatella furcifera (Hemiptera: Delphacidae) identified from next-generation sequence data. J. Insect Sci. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Zhang, W.; Orantes, L.; Jun, T.H.; Mittapalli, O.; Mian, M.A.; Michel, A.P. Combining next-generation sequencing strategies for rapid molecular resource development from an invasive aphid species, Aphis glycines. PLoS ONE 2010, 5, e11370. [Google Scholar] [CrossRef] [PubMed]
- Bouanani, M.A.; Magne, F.; Lecompte, E.; Crouau-Roy, B. Development of 18 novel polymorphic microsatellites from Coccinella septempunctata and cross-species amplification in Coccinellidae species. Conserv. Genet. Resour. 2015, 7, 445–449. [Google Scholar] [CrossRef]
- Meglécz, E.; Nève, G.; Biffin, E.; Gardner, M.G. Breakdown of phylogenetic signal: A survey of microsatellite densities in 454 shotgun sequences from 154 non model Eukaryote species. PLoS ONE 2012, 7, e40861. [Google Scholar] [CrossRef] [PubMed]
- Arunkumar, K.P.; Kifayathullah, L.; Nagaraju, J. Microsatellite markers for the Indian golden silkmoth, Antheraea assama (Saturniidae: Lepidoptera). Mol. Ecol. Resour. 2009, 9, 268–270. [Google Scholar] [CrossRef] [PubMed]
- Dharmarajan, G.; Beatty, W.S.; Rhodes, O.E. Heterozygote deficiencies caused by a Wahlund effect: Dispelling unfounded expectations. J. Wildl. Manag. 2013, 77, 226–234. [Google Scholar] [CrossRef]
- Amsellem, L.; Risterucci, A.M.; Benrey, B. Isolation and characterization of polymorphic microsatellite loci in Lobesia botrana Den. & Schiff. (Lepidoptera: Tortricidae). Mol. Ecol. Notes 2003, 3, 117–119. [Google Scholar]
- An, B.; Deng, X.; Shi, H.; Ding, M.; Lan, J.; Yang, J.; Li, Y. Development and characterization of microsatellite markers for rice leaffolder, Cnaphalocrocis medinalis (Guenee) and cross-species amplification in other Pyralididae. Mol. Biol. Rep. 2014, 41, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Chapuis, M.P.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Coates, B.S.; Sumerford, D.V.; Hellmich, R.L.; Lewis, L.C. Mining an Ostrinia nubilalis midgut expressed sequence tag (EST) library for candidate genes and single nucleotide polymorphisms (SNPs). Insect Mol. Biol. 2008, 17, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Groenewald, C.W.; Born, C.; Chown, S.L.; van Vuuren, B.J. Development of a microsatellite library for the flightless moth Pringleophaga marioni Viette (Lepidoptera: Tineidae). Conserv. Genet. Resour. 2011, 3, 291–294. [Google Scholar] [CrossRef]
- Meglecz, E.; Petenian, F.; Danchin, E.; D’Acier, A.C.; Rasplus, J.Y.; Faure, E. High similarity between flanking regions of different microsatellites detected within each of two species of Lepidoptera: Parnassius apollo and Euphydryas aurinia. Mol. Ecol. 2004, 13, 1693–1700. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.X. Lepidopteran microsatellite DNA: Redundant but promising. Trends Ecol. Evol. 2004, 19, 507–509. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.J.; Wen, J.B.; Wei, S.J.; Liu, J.; Yang, F.; Chen, M. Characterization of novel microsatellite markers for Hyphantria cunea and implications for other Lepidoptera. Bull. Entomol. Res. 2015, 105, 273–284. [Google Scholar] [CrossRef] [PubMed]
- A’Hara, S.; Cottrell, J. Development and characterisation of ten polymorphic microsatellite markers for the pine-tree lappet moth Dendrolimus pini (Lepidoptera: Lasiocampidae). Conserv. Genet. Resour. 2013, 5, 1135–1137. [Google Scholar] [CrossRef]
- Meglécz, E.; Costedoat, C.; Dubut, V.; Gilles, A.; Malausa, T.; Pech, N.; Martin, J.F. QDD: A user-friendly program to select microsatellite markers and design primers from large sequencing projects. Bioinform. 2010, 26, 403–404. [Google Scholar] [CrossRef] [PubMed]
- Blacket, M.J.; Robin, C.; Good, R.T.; Lee, S.F.; Miller, A.D. Universal primers for fluorescent labelling of PCR fragments—an efficient and cost—effective approach to genotyping by fluorescence. Mol. Ecol. Resour. 2012, 12, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Park, S.D.E. Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection. Ph.D. Thesis, University of Dublin, Ireland, UK, 2001. [Google Scholar]
- Antao, T.; Lopes, A.; Lopes, R.J.; Beja-Pereira, A.; Luikart, G. LOSITAN: A workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinform. 2008, 9, 323. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 7, 574–578. [Google Scholar]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
| Repeat Motif | Number of Repeats | Total Frequency (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||
| AC/GT | 7521 | 2765 | 1747 | 1543 | 1433 | 720 | 76 | 2 | 16.612 |
| AG/CT | 2080 | 862 | 512 | 387 | 415 | 340 | 72 | 2 | 4.908 |
| AT/AT | 14,053 | 5758 | 4165 | 4491 | 3380 | 732 | 42 | 2 | 34.285 |
| CG/CG | 1325 | 109 | 12 | 4 | 3 | 3 | 1 | 2 | 1.533 |
| AAC/GTT | 92 | 34 | 18 | 3 | 0.154 | ||||
| AAG/CTT | 2074 | 747 | 142 | 3 | 3.117 | ||||
| AAT/ATT | 8333 | 4004 | 438 | 3 | 13.429 | ||||
| ACC/GGT | 45 | 12 | 6 | 1 | 0.067 | ||||
| ACG/CGT | 457 | 121 | 38 | 3 | 0.651 | ||||
| ACT/AGT | 218 | 71 | 50 | 3 | 0.359 | ||||
| AGC/CTG | 77 | 27 | 14 | 3 | 0.127 | ||||
| AGG/CCT | 49 | 7 | 7 | 1 | 0.067 | ||||
| ATC/ATG | 11,707 | 5017 | 435 | 3 | 18.036 | ||||
| CCG/CGG | 436 | 162 | 93 | 3 | 0.729 | ||||
| AAAC/GTTT | 257 | 4 | 0.274 | ||||||
| AAAG/CTTT | 355 | 4 | 0.377 | ||||||
| AAAT/ATTT | 1798 | 4 | 1.894 | ||||||
| AACC/GGTT | 14 | 4 | 0.019 | ||||||
| AACG/ | 1 | 1 | 0.002 | ||||||
| AACT/AGTT | 42 | 4 | 0.048 | ||||||
| AAGG/CTTC | 8 | 2 | 0.011 | ||||||
| AAGT/ACTT | 62 | 4 | 0.069 | ||||||
| AATC/GATT | 146 | 4 | 0.158 | ||||||
| AATG/CATT | 283 | 4 | 0.302 | ||||||
| AATT/AATT | 68 | 3 | 0.075 | ||||||
| ACAG/CTGT | 112 | 4 | 0.122 | ||||||
| ACAT/ATGT | 1448 | 4 | 1.526 | ||||||
| ACCT/AGGT | 114 | 3 | 0.123 | ||||||
| ACGC/ACGC | 20 | 4 | 0.025 | ||||||
| ACGG/CGTC | 51 | 4 | 0.058 | ||||||
| ACGT/ACGT | 3 | 0.003 | |||||||
| ACTC/GTGA | 20 | 3 | 0.024 | ||||||
| AGAT/ATCT | 473 | 4 | 0.501 | ||||||
| ATCC/ATGG | 216 | 4 | 0.231 | ||||||
| ATGC/ATGC | 6 | 1 | 0.007 | ||||||
| AGGC/CCTG | 10 | 2 | 0.013 | ||||||
| CGAG/CTCG | 2 | 0.002 | |||||||
| CGGC/CGGC | 3 | 0.003 | |||||||
| CTAG/CTAG | 3 | 0.003 | |||||||
| GACT/TCAG | 2 | 1 | 0.003 | ||||||
| GCAA/GCAA | 2 | 0.002 | |||||||
| OTHERS | 46 | 0.048 | |||||||
| DNR | 24,979 | 9494 | 6436 | 6425 | 5231 | 1795 | 191 | 8 | 57.338 |
| TNR | 23,488 | 10,202 | 1241 | 26 | 36.738 | ||||
| TTNR | 5519 | 72 | 5.876 | ||||||
| PNR | 46 | 0.048 | |||||||
| Locus | Dye | Repeat Motif | Primer Sequence (5′–3′) | Allele No. | Size Range (bp) | HWE | r | HO | HE | FIS | PIC | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beijing | Hubei | Beijing | Hubei | Beijing | Hubei | Beijing | Hubei | Beijing | Hubei | |||||||
| CS03 | ROX | (AGT)6 | F: TAAAAGCGATTCGTTGGGAC | 5 | 209–218 | 0.608 | 1.000 | 0.000 | 0.000 | 0.419 | 0.125 | 0.387 | 0.122 | −0.086 | −0.029 | 0.250 |
| R: ATGGCGTCATATCTTCGACC | ||||||||||||||||
| CS04 | FAM | (ACT)6 | F: TTCCGTGCATGTCGTAAGAG | 6 | 120–139 | 0.012 | 0.016 | 0.100 | 0.011 | 0.484 | 0.406 | 0.655 | 0.468 | 0.265 | 0.133 | 0.531 |
| R: CGCGTTTAGCATCAATCTCA | ||||||||||||||||
| CS05 | HEX | (ACG)6 | F: ACACTAGTTGAGTGATTTCAACCG | 5 | 101–113 | 0.000 | 0.000 | 0.323 | 0.269 | 0.097 | 0.188 | 0.622 | 0.631 | 0.847 | 0.706 | 0.572 |
| R: GCATCTGGCTAGATTCTGATGA | ||||||||||||||||
| CS06 | HEX | (CCG)6 | F: ACCGACCAGTCCATTCGAT | 4 | 106–123 | 0.460 | 0.856 | 0.000 | 0.005 | 0.613 | 0.469 | 0.539 | 0.489 | −0.139 | 0.041 | 0.412 |
| R: CTCCTTAGGTCTCTGCGTCG | ||||||||||||||||
| CS07 | HEX | (AAT)6 | F: AGCAGCCTGCATCCAACC | 9 | 99–122 | 0.738 | 0.000 | 0.000 | 0.106 | 0.581 | 0.581 | 0.643 | 0.771 | 0.098 | 0.250 | 0.696 |
| R: ACACACTCCCAATTCGCTTC | ||||||||||||||||
| CS101 | HEX | (AAC)6 | F: TTGGTTCATGGATCTAGGAGG | 4 | 104–115 | 0.007 | 0.006 | 0.145 | 0.132 | 0.161 | 0.219 | 0.309 | 0.354 | 0.483 | 0.385 | 0.304 |
| R: TCCTAAGTCTACCTAACTTTATGTGTT | ||||||||||||||||
| CS102 | FAM | (AGT)6 | F: CCGTAATAATTCGACACAAGCA | 5 | 131–147 | 1.000 | 0.004 | 0.000 | 0.159 | 0.226 | 0.219 | 0.211 | 0.448 | −0.071 | 0.516 | 0.325 |
| R: CCTATACTCGTATACTTAAACAACTGA | ||||||||||||||||
| CS103 | HEX | (AAC)6 | F: AGTATCAAAAGAAACCCCTAA | 4 | 111–120 | 1.000 | 0.700 | 0.011 | 0.036 | 0.355 | 0.594 | 0.373 | 0.661 | 0.049 | 0.104 | 0.506 |
| R: ATCGGCATTATTTGTAAGGT | ||||||||||||||||
| CS11 | HEX | (AAG)6 | F: CCTCGTATTAGATTAGGCGGAA | 4 | 95–112 | 1.000 | 0.000 | 0.000 | 0.200 | 0.065 | 0.250 | 0.063 | 0.560 | −0.017 | 0.558 | 0.343 |
| R: CCCAAGTTGAATGGGAACAG | ||||||||||||||||
| CS14 | HEX | (AGT)6 | F: TGCGACAAAATGCCAGAATA | 6 | 106–136 | 0.020 | 0.952 | 0.129 | 0.000 | 0.355 | 0.594 | 0.590 | 0.554 | 0.403 | −0.074 | 0.489 |
| R: GCCGATGTATTCTAATGAAGCC | ||||||||||||||||
| CS17 | HEX | (AAG)6 | F: CTCAAGAGTTCTATATACGGGG | 5 | 102–117 | 0.000 | 0.001 | 0.294 | 0.170 | 0.233 | 0.219 | 0.751 | 0.448 | 0.693 | 0.516 | 0.592 |
| R: GGCGATGGGATAGCTGTTAC | ||||||||||||||||
| CS18 | HEX | (AAT)6 | F: AGATAGCTCGTTGACAAAGTT | 3 | 111–117 | 0.402 | 0.000 | 0.041 | 0.183 | 0.194 | 0.125 | 0.228 | 0.344 | 0.155 | 0.640 | 0.272 |
| R: TGTTTTGGAAGCAACAAACG | ||||||||||||||||
| CS19 | HEX | (AGT)6 | F: CCAATGTGTCGTACAACGTG | 7 | 113–134 | 0.291 | 0.015 | 0.062 | 0.089 | 0.516 | 0.438 | 0.631 | 0.561 | 0.184 | 0.222 | 0.568 |
| R: CCTCAAGTAAATATAATCAGGGCG | ||||||||||||||||
| CS20 | FAM | (ACT)6 | F: CAAATCCTTGGCAATGTGAA | 4 | 109–126 | 0.030 | 0.000 | 0.076 | 0.224 | 0.462 | 0.156 | 0.646 | 0.496 | 0.290 | 0.688 | 0.478 |
| R: AGAAAAGATTCACCTGCGCT | ||||||||||||||||
| CS21 | FAM | (ACT)6 | F: CGCATTTGCTACTCACCTGT | 4 | 105–120 | 0.000 | 0.000 | 0.201 | 0.248 | 0.000 | 0.063 | 0.178 | 0.383 | 1.000 | 0.839 | 0.256 |
| R: ACTTACATTCACGTTGCCCA | ||||||||||||||||
| CS22 | FAM | (CCG)6 | F: GTAACGAGCGCAATTGATGA | 3 | 122–128 | 0.050 | 1.000 | 0.108 | 0.000 | 0.032 | 0.063 | 0.094 | 0.062 | 0.659 | −0.008 | 0.075 |
| R: CGCGCTAATCTGGTTAATACG | ||||||||||||||||
| CS24 | ROX | (CCG)6 | F: TCTAAGGAGTGTCCGAAGGC | 2 | 247–248 | 1.000 | 1.000 | 0.000 | 0.013 | 0.452 | 0.469 | 0.444 | 0.496 | −0.017 | 0.055 | 0.373 |
| R: TCAAGTACCGTGTGCGGATA | ||||||||||||||||
| CS26 | FAM | (CCG)6 | F: ACCCGAGTAAAGACCCGACT | 4 | 123–135 | 0.000 | 0.105 | 0.272 | 0.097 | 0.129 | 0.065 | 0.535 | 0.182 | 0.762 | 0.649 | 0.360 |
| R: TGTTAACCCTAGAAGGCCCG | ||||||||||||||||
| CS28 | FAM | (ACT)6 | F: GCTGGTGTGGATGGCATAGT | 7 | 126–147 | 0.023 | 0.061 | 0.082 | 0.099 | 0.484 | 0.438 | 0.637 | 0.591 | 0.243 | 0.263 | 0.615 |
| R: AACTTCGAATTTCCATTGCG | ||||||||||||||||
| CS29 | FAM | (ACC)6 | F: TCGGTCACGTTATTTTAGCAA | 9 | 89–147 | 0.000 | 0.000 | 0.173 | 0.266 | 0.290 | 0.290 | 0.504 | 0.525 | 0.428 | 0.451 | 0.494 |
| R: CATGGTCAGTGCTAGGCAGA | ||||||||||||||||
| CS31 | FAM | (ACT)6 | F: CGGACTTCTGAAACCGTGAT | 6 | 129–148 | 0.086 | 0.000 | 0.028 | 0.137 | 0.484 | 0.484 | 0.563 | 0.698 | 0.143 | 0.310 | 0.601 |
| R: GCCAATTCAGTTATGAGGGC | ||||||||||||||||
| CS32 | FAM | (AGG)6 | F: CTAGGTACACCAATCGGCCA | 2 | 134–137 | 0.054 | 0.495 | 0.111 | 0.037 | 0.194 | 0.438 | 0.317 | 0.500 | 0.394 | 0.127 | 0.360 |
| R: GCTGCCATTTCACCAGTCTT | ||||||||||||||||
| CS33 | FAM | (ACT)6 | F: AATAGGGCTCCTCCACACCT | 8 | 130–156 | 0.392 | 0.706 | 0.030 | 0.003 | 0.677 | 0.531 | 0.769 | 0.571 | 0.121 | 0.071 | 0.643 |
| R: GATCTGCAAATCTGCCTGTG | ||||||||||||||||
| CS34 | FAM | (AGT)6 | F: CGCCCTAGACGAACCTACAC | 4 | 130–143 | 0.587 | 1.000 | 0.026 | 0.000 | 0.258 | 0.219 | 0.283 | 0.205 | 0.091 | −0.069 | 0.227 |
| R: GCCTATGTTCAGCAGAAGACG | ||||||||||||||||
| CS35 | ROX | (AAG)6 | F: CAAAGATAATGTACAAAGACGTG | 5 | 113–142 | 0.001 | 0.040 | 0.215 | 0.121 | 0.269 | 0.531 | 0.652 | 0.750 | 0.592 | 0.296 | 0.655 |
| R: CAACTGTCTGCAACACAGCA | ||||||||||||||||
| CS36 | ROX | (CCG)6 | F: CACCGATTTGTTTTATCGCA | 7 | 138–159 | 0.284 | 1.000 | 0.025 | 0.000 | 0.581 | 0.063 | 0.604 | 0.062 | 0.039 | −0.008 | 0.351 |
| R: GGCGCTAATGTCTACCCTCA | ||||||||||||||||
| CS37 | ROX | (ACC)6 | F: TAAGAAGATCCTCGCCCAGA | 2 | 145–148 | 0.159 | 0.300 | 0.081 | 0.000 | 0.097 | 0.406 | 0.151 | 0.329 | 0.362 | −0.240 | 0.215 |
| R: TACATCGTTGTAGGACCGCC | ||||||||||||||||
| CS38 | ROX | (AGC)6 | F: CAAACAAATTATCCGCGTCC | 3 | 147–153 | 0.022 | 0.001 | 0.140 | 0.176 | 0.000 | 0.000 | 0.148 | 0.235 | 1.000 | 1.000 | 0.181 |
| R: GACAGAAACAATAACAACGACGA | ||||||||||||||||
| CS41 | ROX | (AAC)6 | F: CCACTGGGCTATCACTGCTAT | 6 | 140–168 | 0.118 | 0.132 | 0.040 | 0.052 | 0.581 | 0.281 | 0.664 | 0.360 | 0.128 | 0.221 | 0.509 |
| R: TGCAACAGTGACATCACAAGA | ||||||||||||||||
| CS44 | ROX | (AGT)6 | F: AGTGGGCGCCACCTGCAT | 3 | 149–155 | 1.000 | NA | 0.000 | 0.001 | 0.226 | 0.000 | 0.207 | 0.000 | −0.094 | NA | 0.102 |
| R: CCATCTTTGGCTCAGAAAGC | ||||||||||||||||
| CS45 | ROX | (ACT)6 | F: TGGCCGTTATATCATCCACA | 2 | 155–158 | 1.000 | 1.000 | 0.000 | 0.000 | 0.065 | 0.469 | 0.063 | 0.448 | −0.017 | −0.047 | 0.254 |
| R: GGTAGTCCTGGTCAGAGGCA | ||||||||||||||||
| CS47 | ROX | (AGT)7 | F: ACCGGTATTGCTGTATTTGT | 5 | 151–163 | 0.001 | 0.764 | 0.163 | 0.000 | 0.400 | 0.625 | 0.666 | 0.592 | 0.404 | −0.056 | 0.573 |
| R: CAATTTGTGATTAGGTATTTGTTTCAA | ||||||||||||||||
| CS48 | ROX | (AAT)6 | F: TGTAGCAGTCAAGGTCACGG | 3 | 156–162 | 0.048 | 0.000 | 0.071 | 0.219 | 0.484 | 0.194 | 0.666 | 0.497 | 0.277 | 0.614 | 0.556 |
| R: CGCTATAAAAGTGAACGGCG | ||||||||||||||||
| CS53 | ROX | (AAG)6 | F: TCACGTAACCGTCTGGTTCA | 3 | 137–176 | 1.000 | 0.802 | 0.000 | 0.015 | 0.097 | 0.438 | 0.094 | 0.469 | −0.035 | 0.068 | 0.274 |
| R: TCGTCTTTTCTTTCCATCGG | ||||||||||||||||
| CS82 | HEX | (AGT)6 | F: AAAGGCAGATTAACCGACTAGTGT | 2 | 89–106 | 0.000 | 0.000 | 0.293 | 0.198 | 0.000 | 0.000 | 0.389 | 0.173 | 1.000 | 1.000 | 0.247 |
| R: AAATATTTTCGCGTTCATTTCG | ||||||||||||||||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.-Z.; Cao, L.-J.; Zhu, J.-Y.; Wei, S.-J. Development and Characterization of Novel Microsatellite Markers for the Peach Fruit Moth Carposina sasakii (Lepidoptera: Carposinidae) Using Next-Generation Sequencing. Int. J. Mol. Sci. 2016, 17, 362. https://doi.org/10.3390/ijms17030362
Wang Y-Z, Cao L-J, Zhu J-Y, Wei S-J. Development and Characterization of Novel Microsatellite Markers for the Peach Fruit Moth Carposina sasakii (Lepidoptera: Carposinidae) Using Next-Generation Sequencing. International Journal of Molecular Sciences. 2016; 17(3):362. https://doi.org/10.3390/ijms17030362
Chicago/Turabian StyleWang, You-Zhu, Li-Jun Cao, Jia-Ying Zhu, and Shu-Jun Wei. 2016. "Development and Characterization of Novel Microsatellite Markers for the Peach Fruit Moth Carposina sasakii (Lepidoptera: Carposinidae) Using Next-Generation Sequencing" International Journal of Molecular Sciences 17, no. 3: 362. https://doi.org/10.3390/ijms17030362
APA StyleWang, Y.-Z., Cao, L.-J., Zhu, J.-Y., & Wei, S.-J. (2016). Development and Characterization of Novel Microsatellite Markers for the Peach Fruit Moth Carposina sasakii (Lepidoptera: Carposinidae) Using Next-Generation Sequencing. International Journal of Molecular Sciences, 17(3), 362. https://doi.org/10.3390/ijms17030362