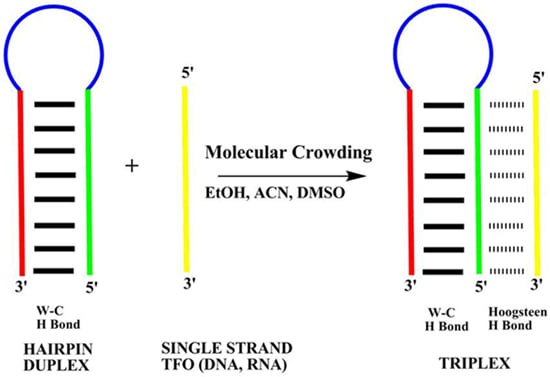
The Effect of Small Cosolutes that Mimic Molecular Crowding Conditions on the Stability of Triplexes Involving Duplex DNA
Abstract
:1. Introduction
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| Hairpin 1 | AGGAAGGAAAAG-(EG)6-CTTTTCCTTCCT |
| DNA TFO 1 | TCCTTCCTTTTC |
| RNA TFO 1 | UCCUUCCUUUUC |
| Hairpin 2 | GGAAAGGAGAAAAGA-(EG)6-TCTTTTCTCCTTTCC |
| DNA TFO 2 | CCTTTCCTCTTTTCT |
| RNA TFO 2 | CCUUUCCUCUUUUCU |
2. Results
2.1. Thermal UV Experiments
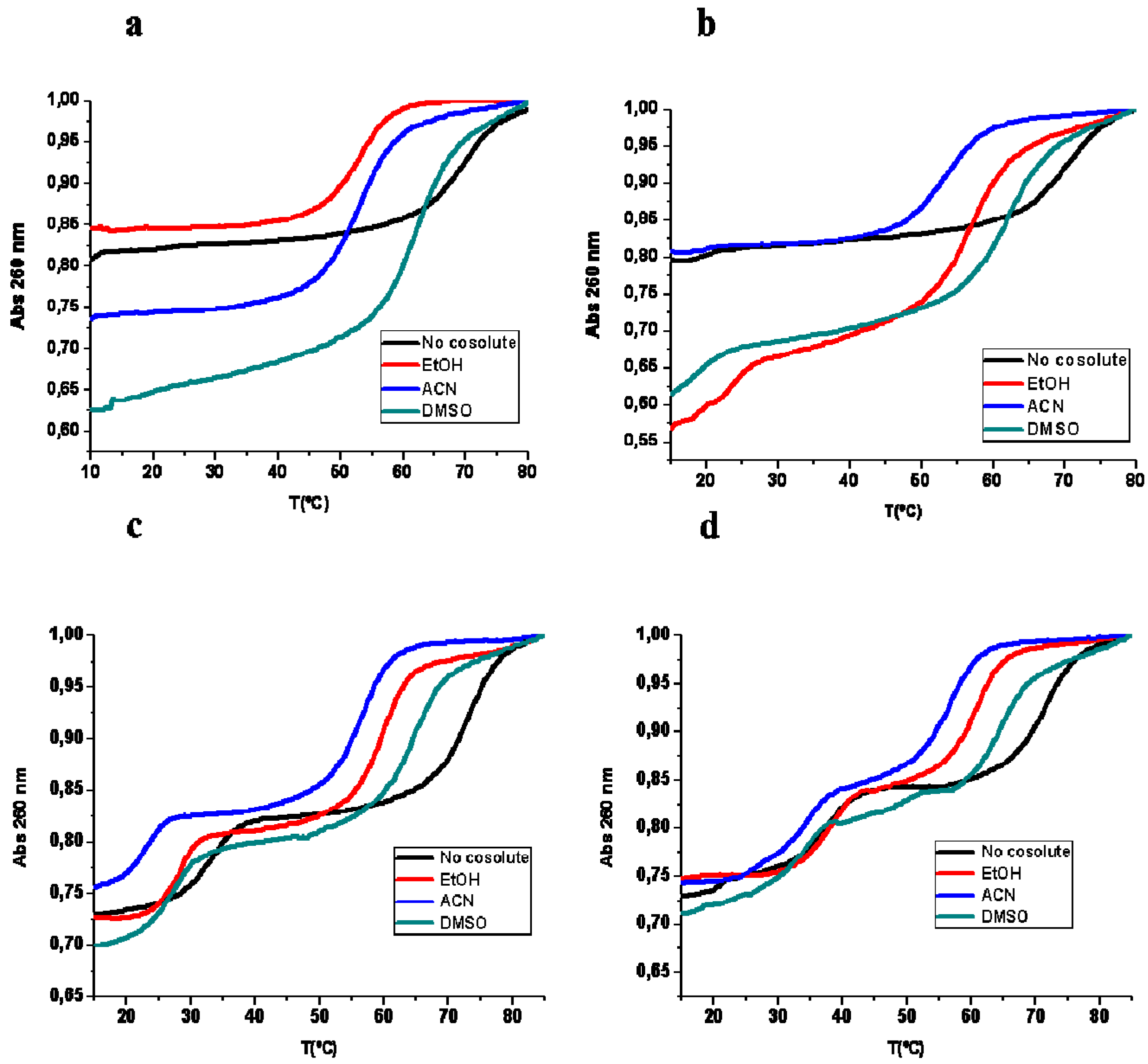
| Tm (°C) | Expected Transition | No Cosolute | 20% EtOH | 20% ACN | 20% DMSO |
|---|---|---|---|---|---|
| Hairpin 1 duplex | Duplex to SS | 67.6 (26) | 50.6 (28) | 53.6 (25) | 61.8 (40) |
| Hairpin 1 duplex + TFO DNA 1 | Triplex to Duplex | 16.0 (3) | – | – | – |
| Duplex to SS | 67.6 (15) | 55.8 (21) | 52.5 (30) | 61.4 (40) | |
| Hairpin 1 duplex + TFO RNA 1 | Triplex to Duplex | 17.9 (2) | 22.7 (17) | 19.2 (2) | 18.5 (11) |
| Duplex to SS | 69.4 (20) | 56.4 (37) | 52.5 (21) | 60.8 (32) | |
| Hairpin 2 duplex | Duplex to SS | 63.0 (29) | 54.6 (29) | 54.7 (30) | 63.4 (40) |
| Hairpin duplex 2 + TFO DNA 2 | Triplex to Duplex | 33.2 (11) | 27.7 (11) | 23.1 (10) | 27.3 (12) |
| Duplex to SS | 72.3 (14) | 59.7 (20) | 56.8 (19) | 64.4 (22) | |
| Hairpin duplex 2 + TFO RNA 2 | Triplex to Duplex | 37.6 (13) | 37.0 (14) | 31.8 (13) | 33.4 (9) |
| Duplex to SS | 71.5 (19) | 61.0 (17) | 56.9 (17) | 64.7 (18) |
| ΔG°37 (Kcal/mol) | Expected Transition | No Cosolute | 20% EtOH | 20% ACN | 20% DMSO |
|---|---|---|---|---|---|
| Hairpin 1 | Duplex to SS | −6.3 | −3.4 | −4.0 | −5.2 |
| Hairpin 1 + TFO DNA 1 | Triplex to Duplex | 0.8 | – | – | – |
| Duplex to SS | −6.5 | −4.6 | −3.5 | −5.0 | |
| Hairpin 1 + TFO RNA 1 | Triplex to Duplex | 1 | −2.6 | 1.3 | −1.5 |
| Duplex to SS | −6.9 | −4.7 | −3.4 | −5.2 | |
| Hairpin 2 | Duplex to SS | −5.9 | −4.3 | −3.7 | −5.5 |
| Hairpin 2 + TFO DNA 1 | Triplex to Duplex | −7.2 | −3.7 | −0.6 | −4.3 |
| Duplex to SS | −9.4 | −7.8 | −5.9 | −8.6 | |
| Hairpin 2 + TFO RNA 1 | Triplex to Duplex | −9.2 | −8.9 | −7.5 | −7.6 |
| Duplex to SS | −11.2 | −8.3 | −6.2 | −10.0 |
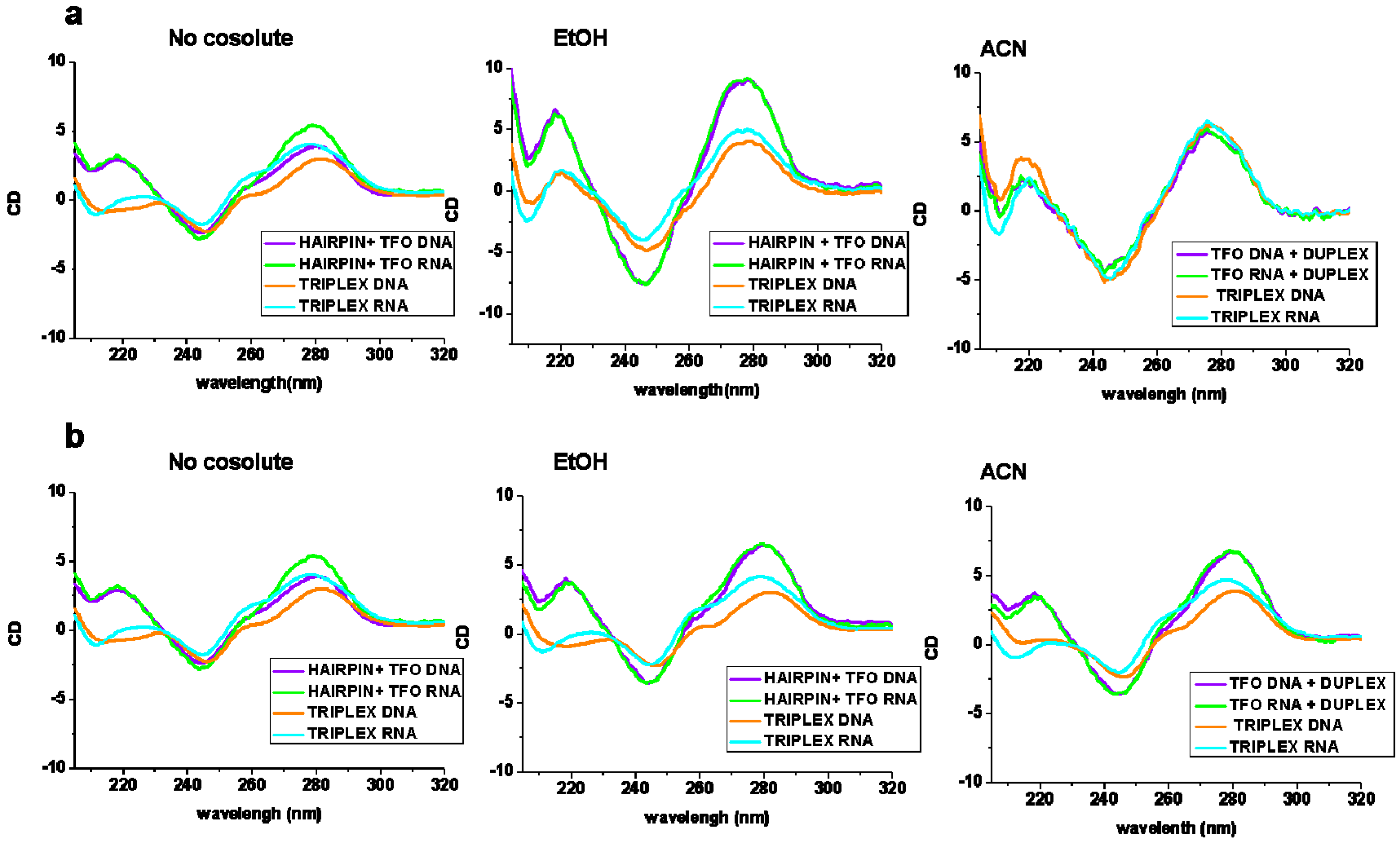
2.2. Circular Dichroism Experiments
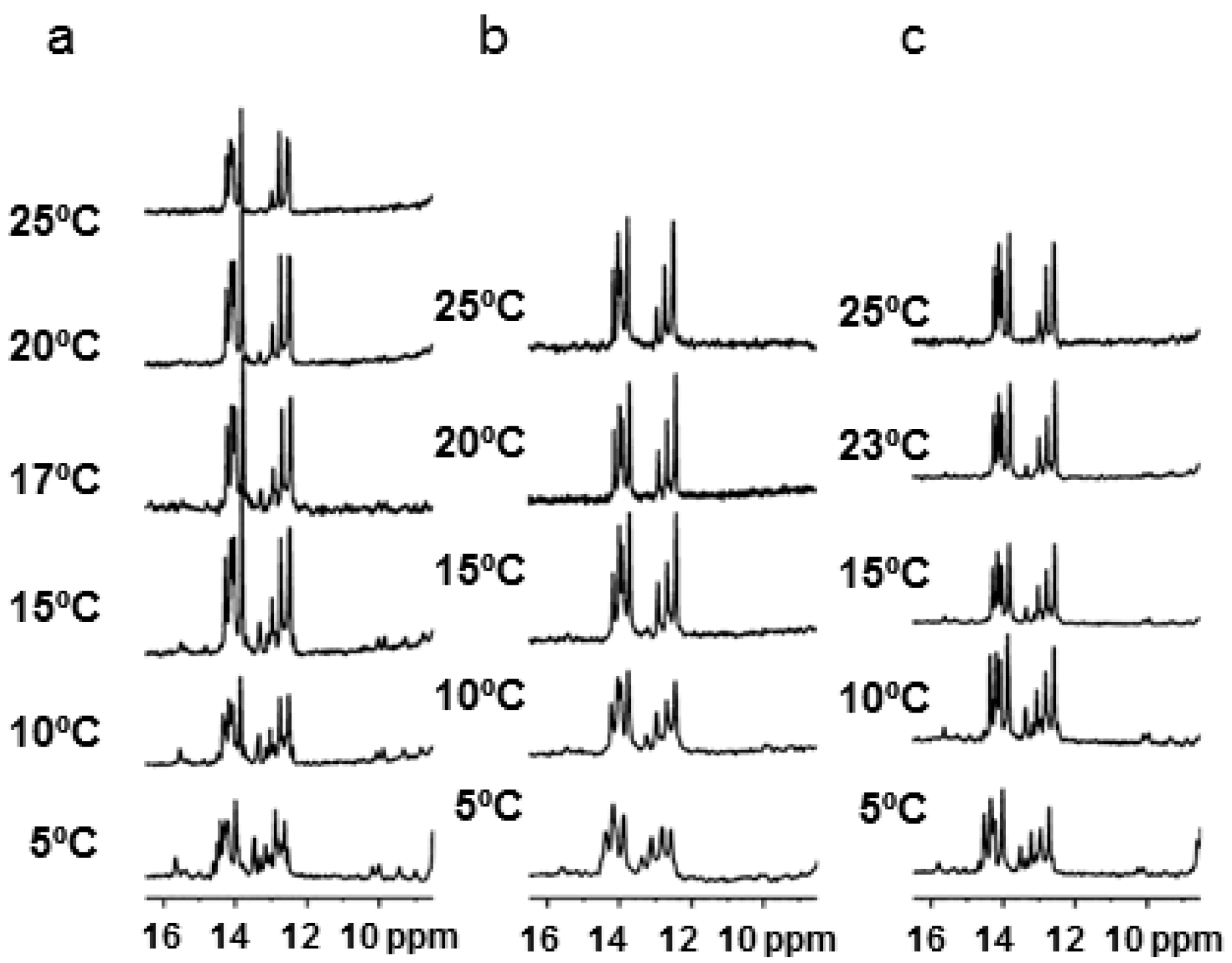
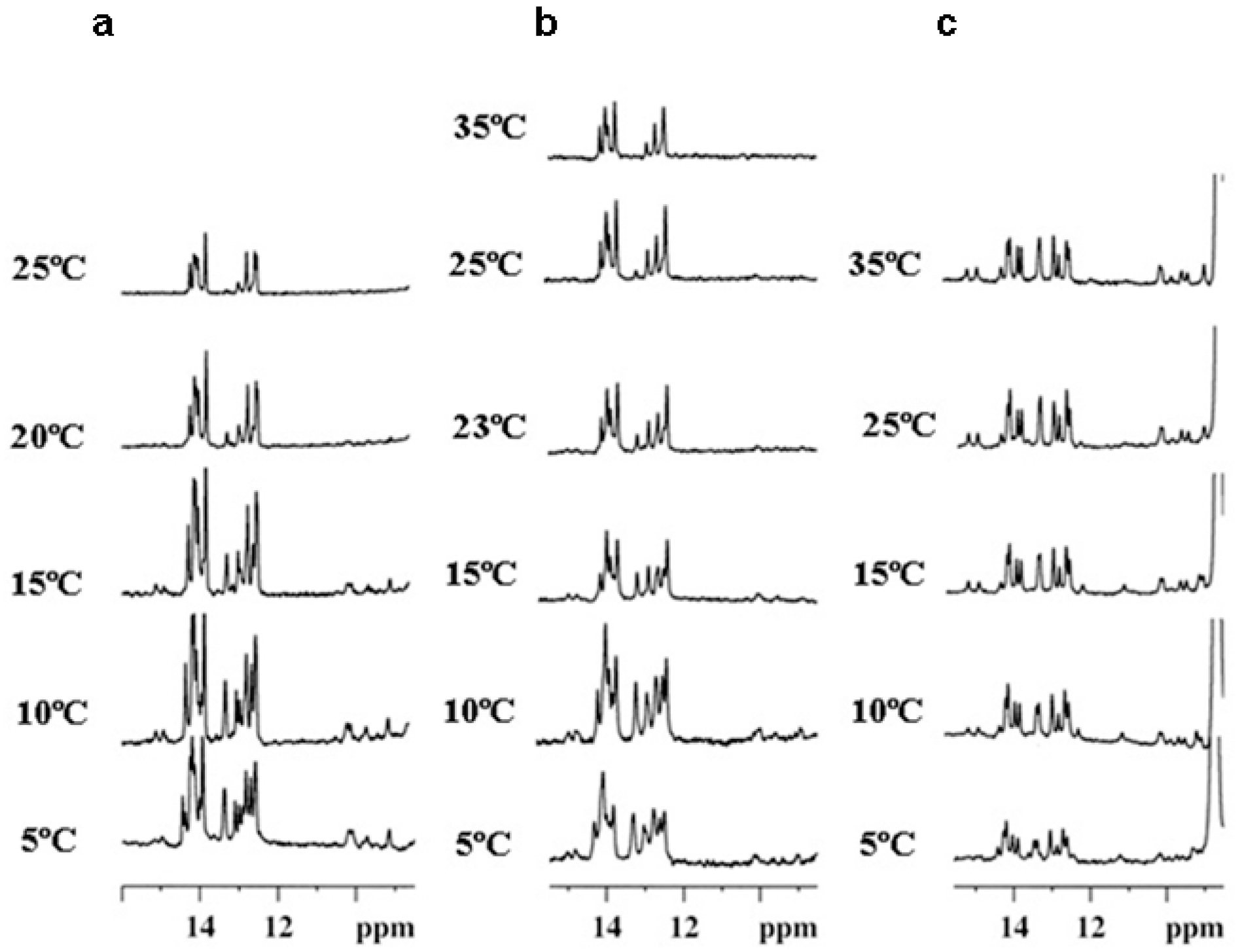
2.3. NMR Studies
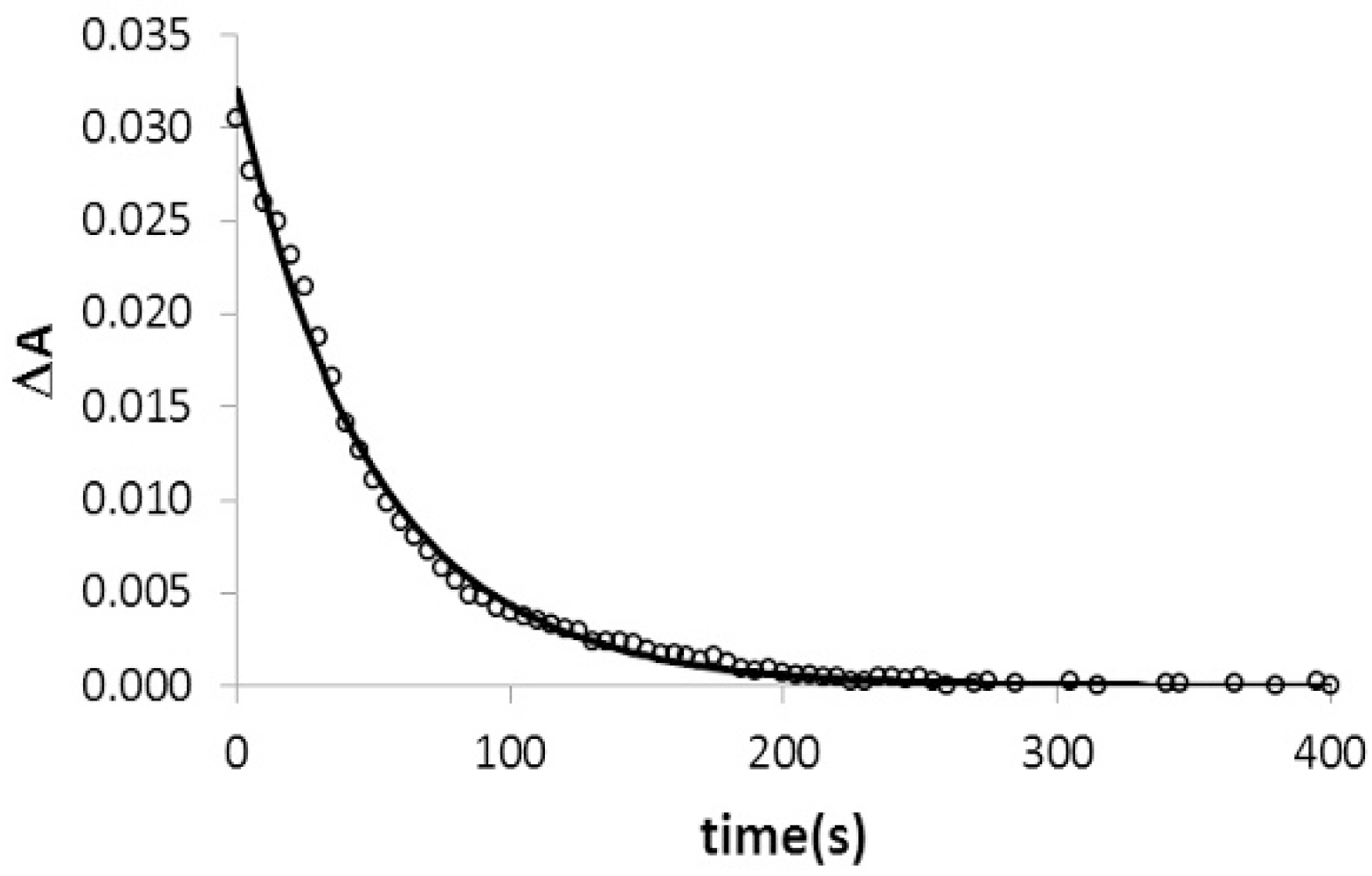
2.4. Kinetic Studies of Triplex Formation under Crowding Conditions
2.5. Effect of Cosolutes on Cation Binding to Triplex Structure
| Tm (°C) | Expected Transition | 10 mM NaCl No Cosolute | 10 mM NaCl 20% EtOH | 1000 mM NaCl No Cosolute | 1000 mM NaCl 20% EtOH |
|---|---|---|---|---|---|
| Hairpin duplex 1 | Duplex to SS | 58.7 | 57.0 | >80 | 62.1 |
| Hairpin duplex 1 + TFO DNA 1 | Triplex to Duplex | – | – | 23 | – |
| Duplex to SS | 58.6 | 57.5 | >80 | 61.8 | |
| Hairpin duplex 1 + TFO RNA 1 | Triplex to Duplex | – | – | 23 | 18.0 |
| Duplex to SS | 57.5 | 54 | >80 | 61.6 |
3. Discussion
4. Experimental Section
4.1. Oligonucleotide Synthesis and Purification
4.2. UV Absorbance and Circular Dichroism Measurements
4.3. NMR Spectroscopy
4.4. TFO Association Analysis by Absorbance Decay
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bochman, M.L.; Katrin Paeschke, K.; Zakian, V.A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Sainia, N.; Zhanga, Y.; Usdinb, K.; Lobacheva, K.S. When secondary comes first—The importance of non-canonical DNA structures. Biochimie 2013, 95, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Glazer, P.M. Triplex DNA: Fundamentals, advances, and potential applications for gene therapy. J. Mol. Med. 1997, 75, 267–282. [Google Scholar] [CrossRef] [PubMed]
- Praseuth, D.; Guieysse, A.L.; Hélène, C. Triple helix formation and the antigene strategy for sequence-specific control of gene expression. Biochim. Biophys. Acta 1999, 1489, 181–206. [Google Scholar] [CrossRef]
- Mirkin, S.M.; Lyamichev, V.I.; Drushlyak, K.N.; Dobrynin, V.M.; Filippov, S.A.; Frank-Kamenetskii, M.D. DNA H form requires a homopurine-homopyrimidine mirror repeat. Nature 1987, 330, 495–497. [Google Scholar] [CrossRef] [PubMed]
- LeDoan, T.; Perrouault, L.; Praseuth, D.; Habhoub, N.; Decout, J.L.; Thuong, N.T.; L’homme, J.; Hélène, C. Sequence-specific recognition, photocrosslinking and cleavage of the DNA double helix by an oligo-(α)-thymidylate covalently linked to an azidoproflavine derivative. Nucleic Acids Res. 1987, 15, 7749–7760. [Google Scholar] [CrossRef]
- Moser, H.E.; Dervan, P.B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science 1987, 238, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Vasquez, K.M.; Wilson, J.H. Triplex-directed modification of genes and gene activity. Trends Biochem. Sci. 1998, 23, 4–9. [Google Scholar] [CrossRef]
- Vasquez, K.M.; Glazer, P.M. Triplex-forming oligonucleotides: principles and applications. Q. Rev. Biophys. 2002, 35, 89–107. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, I.; Patel, D.J. DNA triplexes: Solution structures, hydration sites, energetics, interactions, and function. Biochemistry 1994, 33, 11405–11416. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.W.; Crothers, D.M. Stability and properties of double and triple helices: Dramatic effects of RNA or DNA backbone composition. Science 1992, 258, 1463–1466. [Google Scholar] [CrossRef] [PubMed]
- Escudé, C.; Sun, J.S.; Rougée, M.; Garestier, T.; Hélène, C. Stable triple helices are formed upon binding of RNA oligonucleotides and their 2′-O-methyl derivatives to double-helical DNA. C. R. Acad. Sci. III 1992, 315, 521–525. [Google Scholar] [PubMed]
- Escudé, C.; Francois, J.C.; Sun, J.S.; Ott, G.; Sprinzl, M.; Garestier, T.; Hélène, C. Stability of triple helices containing RNA and DNA strands: experimental and molecular modeling studies. Nucleic Acids Res. 1993, 21, 5547–5553. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Dervan, P.B. Sequence-specific recognition of double helical RNA and RNA.DNA by triple helix formation. Proc. Natl. Acad. Sci. USA 1993, 90, 3806–3810. [Google Scholar] [CrossRef] [PubMed]
- Gotfredsen, C.H.; Schultze, P.; Feigon, J. Solution structure of an intramolecular pyrimidine-purine-pyrimidine triplex containing an RNA third strand. J. Am. Chem. Soc. 1998, 120, 4281–4289. [Google Scholar] [CrossRef]
- Nakano, S.; Miyoshi, D.; Sugimoto, N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem. Rev. 2014, 114, 2733–2758. [Google Scholar] [CrossRef] [PubMed]
- Tateishi-Karimta, H.; Sugimoto, N. Control of stability and structure of nucleic acids using cosolutes. Methods 2014, 67, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, D.; Sugimoto, N. Molecular crowding effects on structure and stability of DNA. Biochimie 2008, 90, 1040–1051. [Google Scholar]
- Petraccone, L.; Pagano, B.; Giancola, C. Studying the effect of crowding and dehydration on DNA G-quadruplexes. Methods 2012, 57, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Albergo, D.D.; Turner, D.H. Solvent effects on the thermodynamics of double-helix formation in (dG-dC)3. Biochemistry 1981, 20, 1413–1418. [Google Scholar] [CrossRef] [PubMed]
- Hickey, D.R.; Turner, D.H. Solvent effects on the stability of A7U7p. Biochemistry 1985, 24, 2086–2094. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Yamaguchi, D.; Tateishi-Karimata, H.; Miyoshi, D.; Sugimoto, N. Hydration changes upon DNA folding studied by osmotic stress experiments. Biophys. J. 2012, 102, 2808–2817. [Google Scholar] [CrossRef] [PubMed]
- Spink, C.H.; Chaires, J.B. Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry 1999, 38, 496–509. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Karimata, H.; Ohmichi, T.; Kawakami, J.; Sugimoto, N. The effect of molecular crowding with nucleotide length and cosolute structure on DNA duplex stability. J. Am. Chem. Soc. 2004, 126, 14330–14331. [Google Scholar] [CrossRef] [PubMed]
- Rozners, E.; Moulder, J. Hydration of short DNA, RNA and 2′-OMe oligonucleotides determined by osmotic stressing. Nucleic Acids Res. 2004, 32, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, D.; Nakamura, K.; Tateishi-Karimata, H.; Ohmichi, T.; Sugimoto, N. Hydration of Watson-Crick base pairs and dehydration of Hoogsteen base pairs inducing structural polymorphism under molecular crowding conditions. J. Am. Chem. Soc. 2009, 131, 3522–3531. [Google Scholar] [CrossRef] [PubMed]
- Collie, G.W.; Parkinson, G.N. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011, 40, 5867–5892. [Google Scholar] [CrossRef] [PubMed]
- Heddi, B.; Phan, A.T. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011, 133, 9824–9833. [Google Scholar] [CrossRef] [PubMed]
- Vorlíčková, M.; Bednářová, K.; Kejnovská, I.; Kypr, J. Intramolecular and intermolecular guanine quadruplexes of DNA in aqueous salt and ethanol solutions. Biopolymers 2007, 26, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.X.; Cui, Y.; Zhao, T.; Fu, H.W.; Koirala, D.; Punnoose, J.A.; Kong, D.M.; Mao, H. Divalent cations and molecular crowding buffers stabilize G-triplex at physiologically relevant temperatures. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Nagatoishi, S.; Isono, N.; Tsumoto, K.; Sugimoto, N. Hydration is required in DNA G-quadruplex-protein binding. ChemBioChem 2011, 12, 1822–1826. [Google Scholar] [CrossRef] [PubMed]
- Spink, C.H.; Chaires, J.B. Selective stabilization of triplex DNA by Poly(ethylene glycols). J. Am. Chem. Soc. 1995, 117, 12887–12888. [Google Scholar] [CrossRef]
- Goobes, R.; Cohen, O.; Minsky, A. Unique condensation patterns of triplex DNA: Physical aspects and physiological implications. Nucleic Acids Res. 2002, 30, 2154–2161. [Google Scholar] [CrossRef] [PubMed]
- Goobes, R.; Minsky, A. Thermodynamic aspects of triplex DNA formation in crowded environments. J. Am. Chem. Soc. 2001, 123, 12692–12693. [Google Scholar] [CrossRef] [PubMed]
- Mamajanov, I.; Engelhart, A.E.; Bean, H.B.; Hud, N.V. DNA and RNA in anhydrous media: Duplex, triplex, and G-quadruplex secondary structures in a deep eutectic solvent. Angew. Chem. Int. Ed. 2010, 49, 6310–6314. [Google Scholar] [CrossRef] [PubMed]
- Schwarz-Finsterle, J.; Stein, S.; Grossmann, C.; Schmitt, E.; Trakhtenbrot, L.; Rechavi, G.; Amariglio, N.; Cremer, C.; Hasmann, M. Comparison of triple helical COMBO-FISH and standard FISH by means of quantitative microscopic image analysis. J. Biochem. Biophys Methods 2007, 70, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Friedman, A.E.; Kool, E.T. Origins of high sequence selectivity: A stopped-flow kinetics study of DNA/RNA hybridization by duplex- and triplex-forming oligonucleotides. Biochemistry 1995, 34, 9774–9784. [Google Scholar] [CrossRef] [PubMed]
- Bernal-Méndez, E.; Leumann, C.J. Stability and kinetics of nucleic acid triplexes with chimaeric DNA/RNA third strands. Biochemistry 2002, 41, 12343–12349. [Google Scholar] [CrossRef] [PubMed]
- Puglisi, J.D.; Tinoco, I. Absorbance melting curves of RNA. Methods Enzymol. 1989, 180, 304–325. [Google Scholar] [PubMed]
- Gowers, D.M.; Fox, K.R. Towards mixed sequence recognition by triple helix formation. Nucleic Acids Res. 1999, 27, 1569–1577. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, N.; Wu, P.; Hara, H.; Kawamoto, Y. pH and cation effects on the properties of parallel pyrimidine motif DNA triplexes. Biochemistry 2001, 40, 9396–9405. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Miles, H.T.; Frazier, J.; Sasisekharan, V. A novel DNA duplex. A parallel-stranded DNA helix with Hoogsteen base pairing. Biochemistry 1993, 32, 11802–11809. [Google Scholar] [CrossRef] [PubMed]
- Xodo, L.E. Kinetic analysis of triple-helix formation by pyrimidine oligodeoxynucleotides and duplex DNA. Eur. J. Biochem. 1995, 228, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Torigoe, H.; Ryuji Shimizume, R.; Sarai, A.; Shindo, H. Triplex formation of chemically modified homopyrimidine oligonucleotides: Thermodynamic and kinetic studies. Biochemistry 1999, 38, 14653–14659. [Google Scholar] [CrossRef] [PubMed]
- Rougée, M.; Faucon, B.; Mergny, J.L.; Barcelo, F.; Giovannangeli, C.; Garestier, T.; Hélène, C. Kinetics and thermodynamics of triple-helix formation: Effects of ionic strength and mismatched. Biochemistry 1992, 31, 9269–9278. [Google Scholar]
- Hari, Y.; Ijitsu, S.; Akabane-Nakata, M.; Yoshida, T.; Obika, S. Kinetic study of the binding of triplex-forming oligonucleotides containing partial cationic modifications to double-stranded DNA. Bioorg. Med. Chem. Lett. 2014, 24, 3046–3049. [Google Scholar] [CrossRef] [PubMed]
- Puri, N.; Majumdar, A.; Cuenoud, B.; Miller, P.S.; Seidman, M.M. Importance of clustered 2′-O-(2-aminoethyl) residues for the gene targeting activity of triple helix-forming oligonucleotides. Biochemistry 2004, 43, 1343–1351. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.J.; van Dyke, M.W. Monovalent cation effects on intermolecular purine-purine-pyrimidine triple-helix formation. Nucleic Acids Res. 1993, 21, 5630–5635. [Google Scholar] [CrossRef] [PubMed]
- Sekharudua, C.Y.; Yathindraa, N.; Sundaralingama, M. Molecular dynamics investigations of DNA triple helical models: Unique features of the Watson-Crick duplex. J. Biomol. Struct. Dyn. 1993, 11, 225–244. [Google Scholar] [CrossRef] [PubMed]
- Alberti, P.; Arimondo, P.P.; Mergny, J.L.; Garestier, T.; Hélène, C.; Sun, J.S. A directional nucleation-zipping mechanism for triple helix formation. Nucleic Acids Res. 2002, 30, 5407–5415. [Google Scholar]
- Hansel, R.; Lohr, F.; Foldynova-Trantirkova, S.; Bamberg, E.; Trantirek, L.; Dotsch, V. The parallel G-quadruplex structure of vertebrate telomeric repeat sequences is not the preferred folding topology under physiological conditions. Nucleic Acids Res. 2011, 39, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Goñi, J.R.; de la Cruz, X.; Orozco, M. Triplex-forming oligonucleotide target sequences in the human genome. Nucleic Acids Res. 2004, 32, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Bagasra, O.; Stir, A.E.; Pirisi-Creek, L.; Creek, K.E.; Bagasra, A.U.; Glenn, N.; Lee, J.S. Role of micro-RNAs in regulation of lentiviral latency and persistence. Appl. Immunohistochem. Mol. Morphol. 2006, 14, 276–290. [Google Scholar] [CrossRef]
- Kanak, M.; Alseiari, M.; Balasubramanian, P.; Addanki, K.; Aggarwal, M.; Noorali, S.; Kalsum, A.; Mahalingam, K.; Pace, G.; Panasik, N.; et al. Triplex-forming microRNAs form stable complexes with HIV-1 provirus and inhibit its replication. Appl. Immunohistochem. Mol. Morphol. 2010, 18, 532–545. [Google Scholar] [CrossRef] [PubMed]
- Toscano-Garibay, J.D.; Aquino-Jarquin, G. Transcriptional regulation mechanism mediated by miRNA-DNA·DNA triplex structure stabilized by Argonaute. Biochim. Biophys. Acta 2014, 1839, 1079–1083. [Google Scholar] [CrossRef] [PubMed]
- Buske, F.A.; Mattick, J.S.; Bailey, T.L. Potential in vivo roles of nucleic acid triple-helices. RNA Biol. 2011, 8, 427–439. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aviñó, A.; Mazzini, S.; Gargallo, R.; Eritja, R. The Effect of Small Cosolutes that Mimic Molecular Crowding Conditions on the Stability of Triplexes Involving Duplex DNA. Int. J. Mol. Sci. 2016, 17, 211. https://doi.org/10.3390/ijms17020211
Aviñó A, Mazzini S, Gargallo R, Eritja R. The Effect of Small Cosolutes that Mimic Molecular Crowding Conditions on the Stability of Triplexes Involving Duplex DNA. International Journal of Molecular Sciences. 2016; 17(2):211. https://doi.org/10.3390/ijms17020211
Chicago/Turabian StyleAviñó, Anna, Stefania Mazzini, Raimundo Gargallo, and Ramon Eritja. 2016. "The Effect of Small Cosolutes that Mimic Molecular Crowding Conditions on the Stability of Triplexes Involving Duplex DNA" International Journal of Molecular Sciences 17, no. 2: 211. https://doi.org/10.3390/ijms17020211