Salivary Microbiome Diversity in Caries-Free and Caries-Affected Children
Abstract
:1. Introduction
2. Results
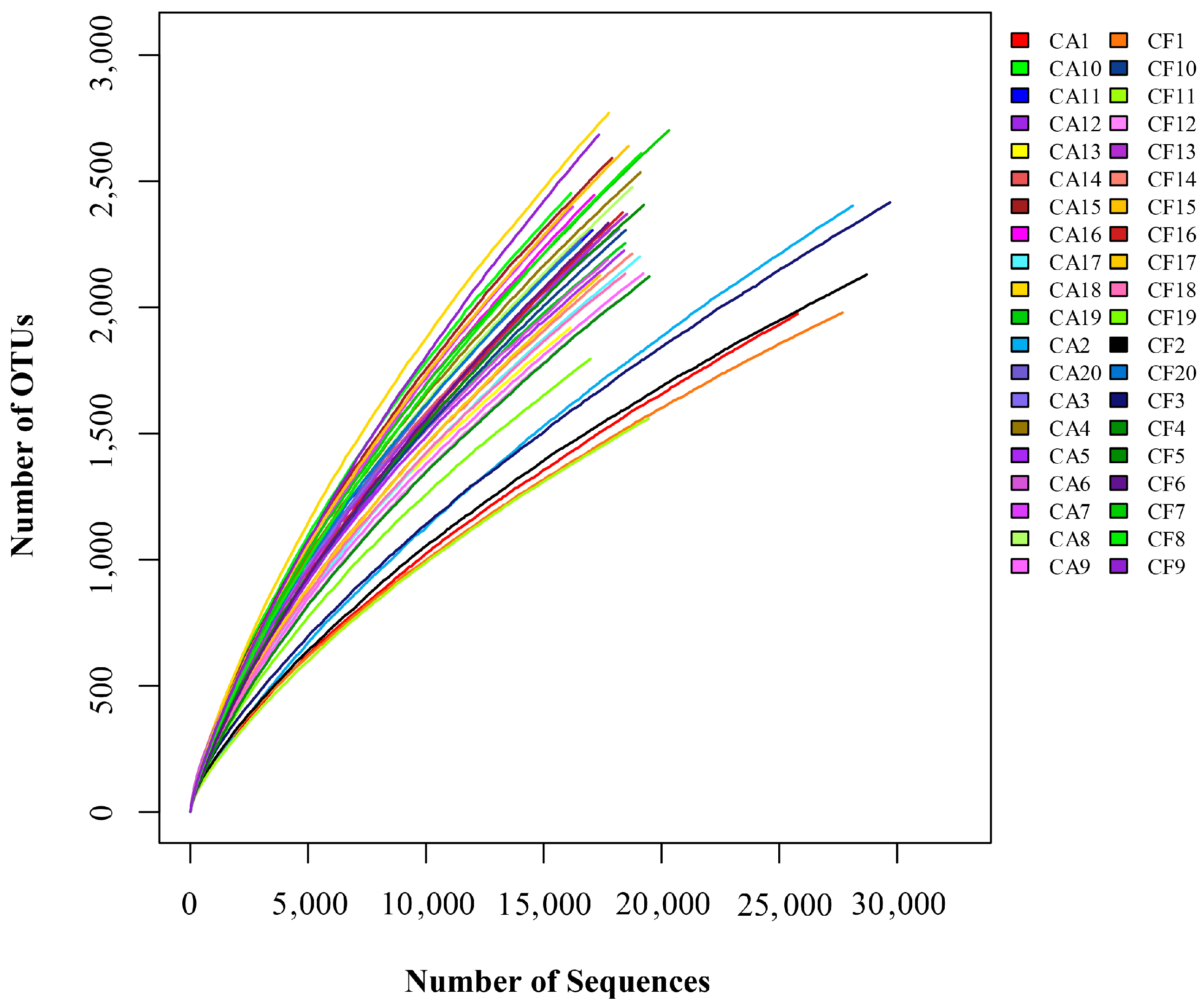
2.1. Features of Salivary Microbiomes
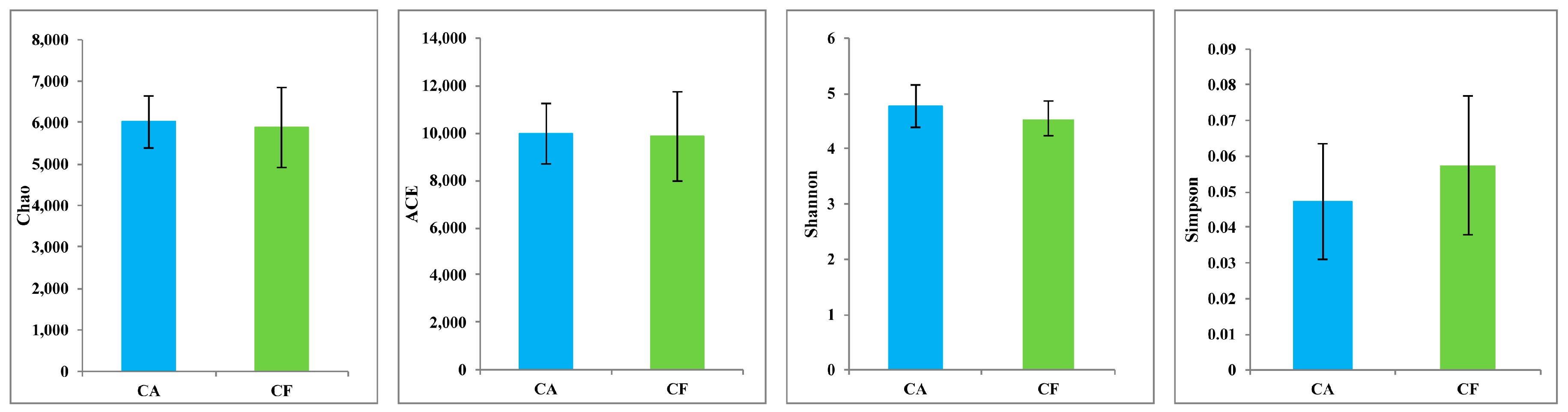
2.2. Microbiome Diversity and Richness
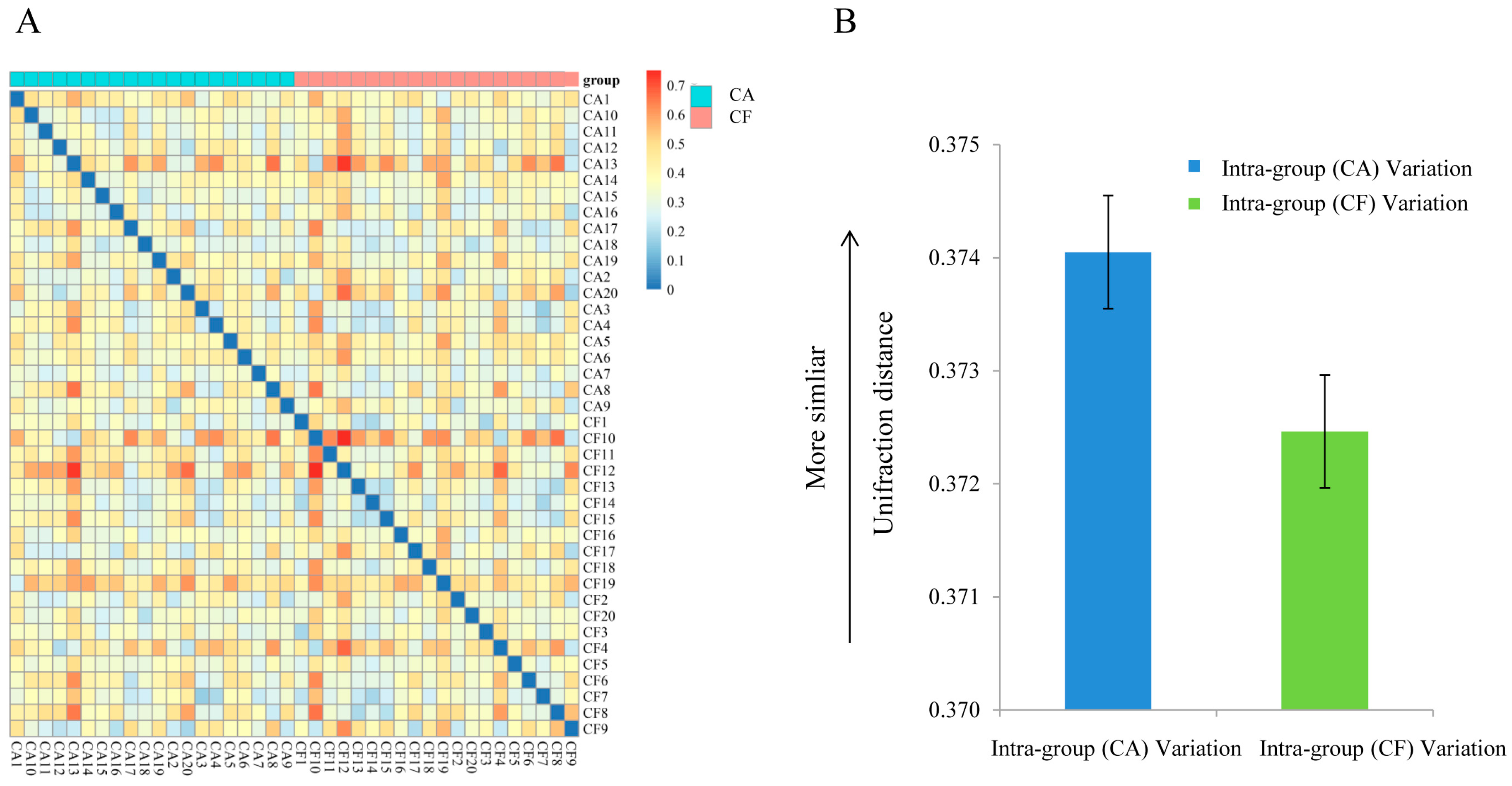
2.3. Community Structures
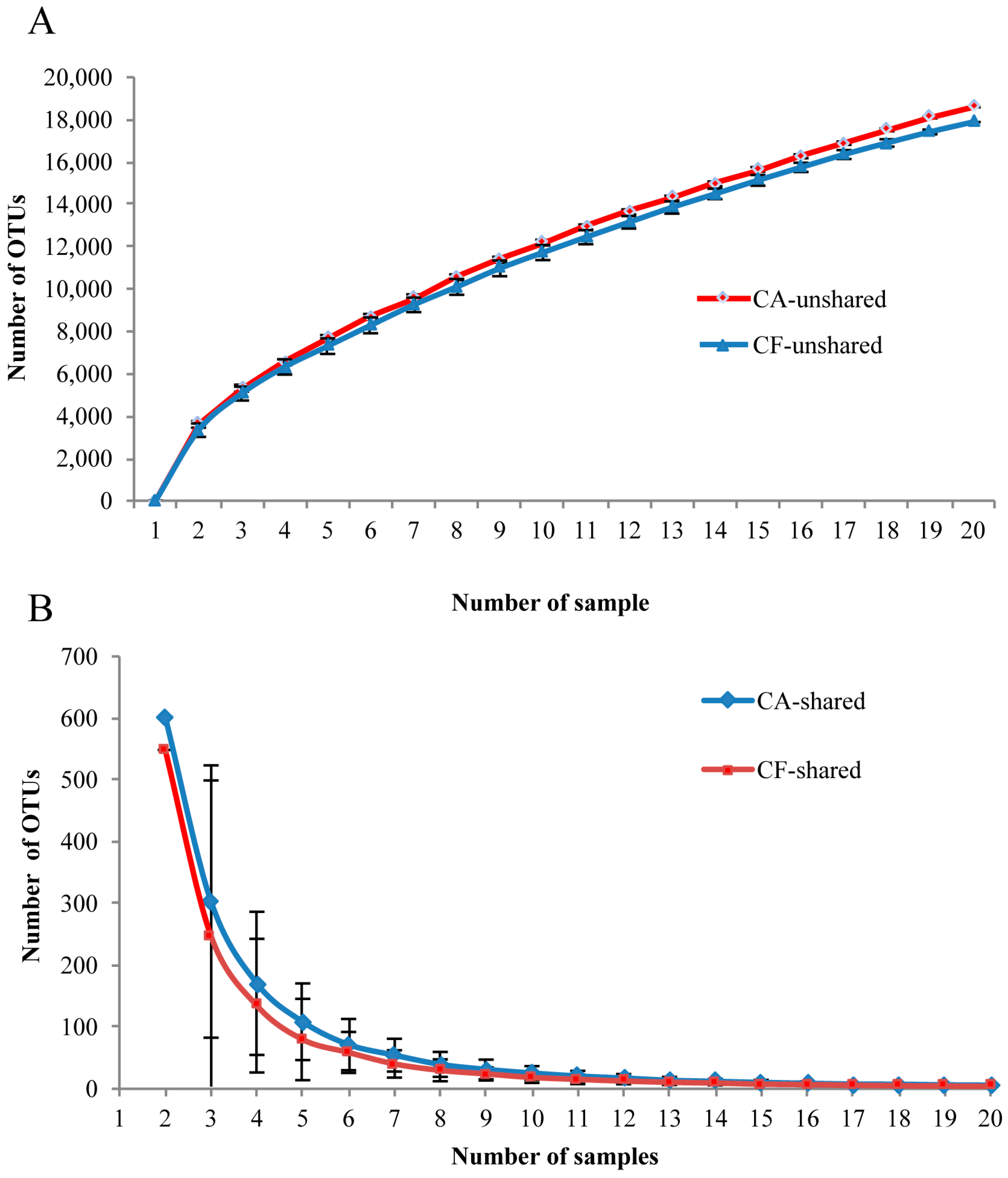
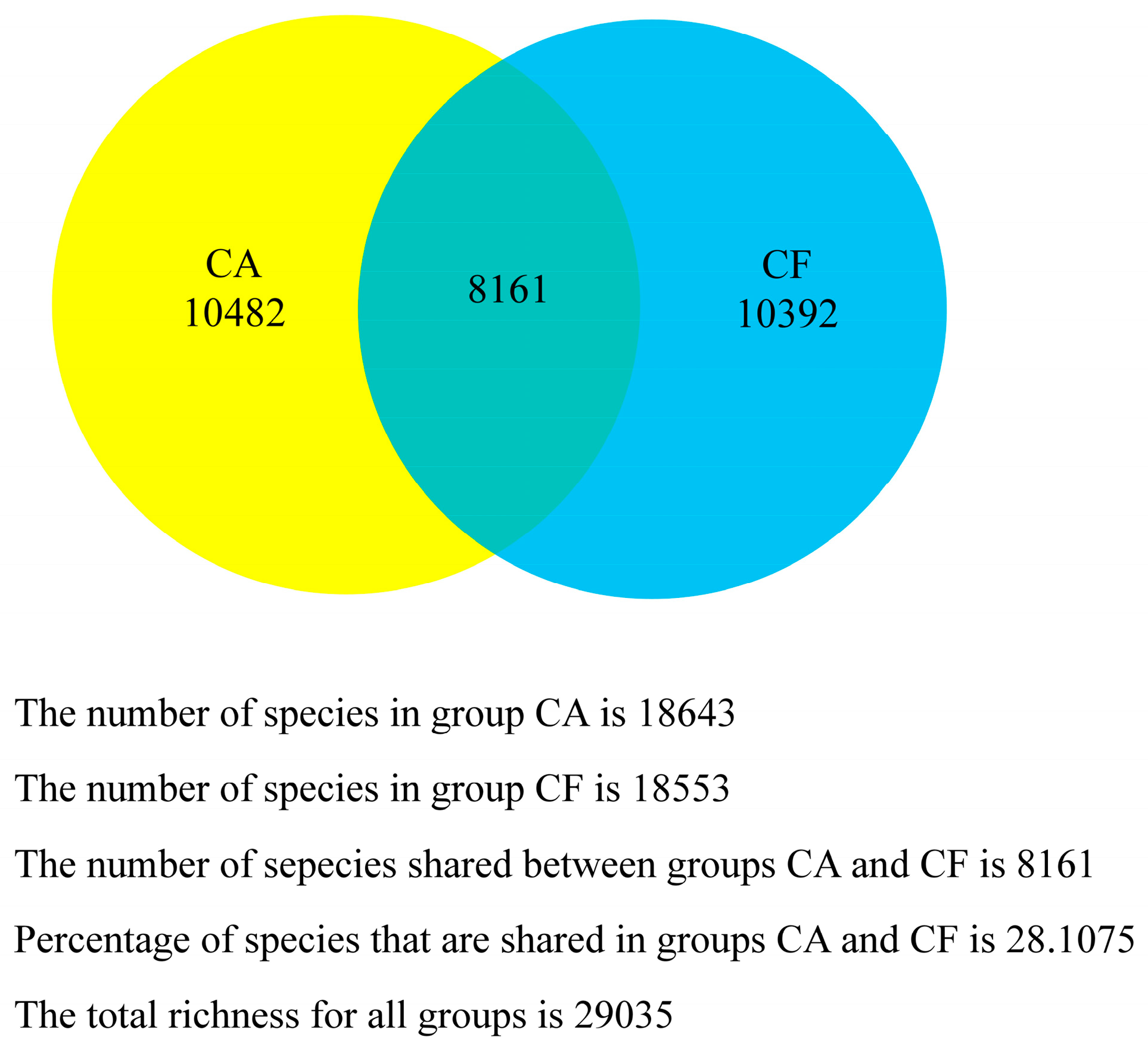
2.4. Core Salivary Microbiome
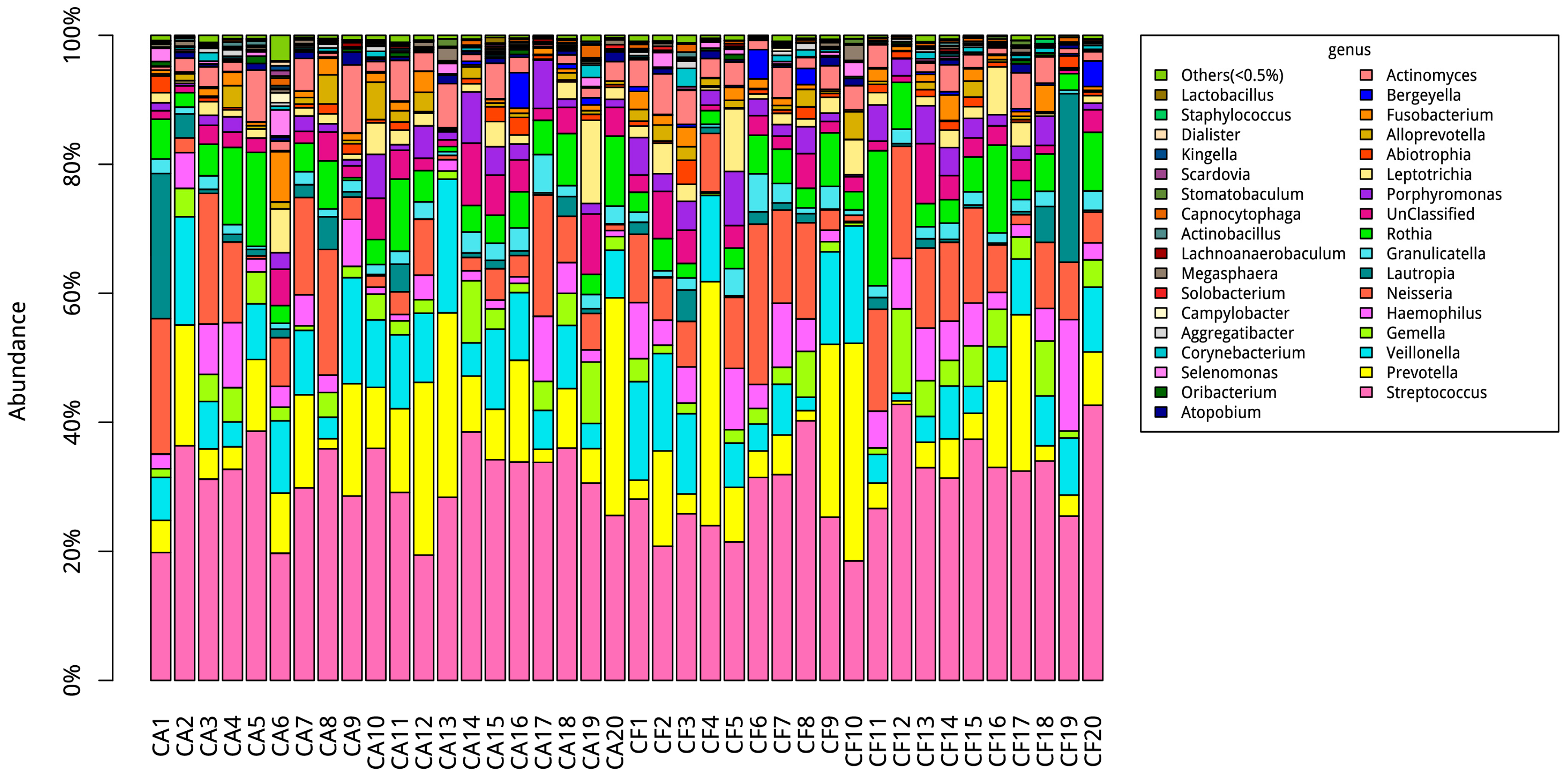
2.5. Comparison of Bacterial Composition between the Caries-Affected and Caries-Free Groups
3. Discussion
4. Materials and Methods
4.1. Sample Size Calculation
4.2. Subject Recruitment and Oral Examination
4.3. Saliva Collection
4.4. DNA Extraction
4.5. PCR Amplification of 16S rRNA Genes and Miseq Sequencing
4.6. 16S rRNA Data Analysis
4.7. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ACE | Abundance-based coverage estimator |
| AW | Aka Ethanol Wash |
| AE | Aka Ethanol |
| CA | Caries-affected |
| CF | Caries-free |
| dmft | Decayed, missing and filled teeth |
| HOMIM | Human Oral Microbe Identification Microarray |
| LED | Light-emitting diode |
| OTU | Operational taxonomic units |
| PCR-DGGE | Polymerase chain reaction denaturing gradient gel electrophoresis |
References
- World Health Organization. Oral Health. Fact Sheet No 318. 2012. Available online: www.who.int/mediacentre/factsheets/fs318/en/index.htm (accessed on 1 June 2016).
- Casamassimo, P.S.; Thikkurissy, S.; Edelstein, B.L.; Maiorini, E. Beyond the dmft: The human and economic cost of early childhood caries. J. Am. Dent. Assoc. 2009, 140, 650–657. [Google Scholar] [CrossRef] [PubMed]
- Marsh, P.D. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 1994, 8, 263–271. [Google Scholar] [PubMed]
- Ajdic, D.; McShan, W.M.; McLaughlin, R.E.; Savic, G.; Chang, J.; Carson, M.B.; Primeaux, C.; Tian, R.; Kenton, S.; Jia, H.; et al. Genome sequence of streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 2002, 99, 14434–14439. [Google Scholar] [CrossRef] [PubMed]
- Corby, P.M.; Lyons-Weiler, J.; Bretz, W.A.; Hart, T.C.; Aas, J.A.; Boumenna, T.; Goss, J.; Corby, A.L.; Junior, H.M.; Weyant, R.J.; et al. Microbial risk indicators of early childhood caries. J. Clin. Microbiol. 2005, 43, 5753–5759. [Google Scholar] [CrossRef] [PubMed]
- Marchant, S.; Brailsford, S.R.; Twomey, A.C.; Roberts, G.J.; Beighton, D. The predominant microflora of nursing caries lesions. Caries Res. 2001, 35, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Klinke, T.; Kneist, S.; de Soet, J.J.; Kuhlisch, E.; Mauersberger, S.; Forster, A.; Klimm, W. Acid production by oral strains of candida albicans and lactobacilli. Caries Res. 2009, 43, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Raja, M.; Hannan, A.; Ali, K. Association of oral candidal carriage with dental caries in children. Caries Res. 2010, 44, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Luo, A.H.; Yang, D.Q.; Xin, B.C.; Paster, B.J.; Qin, J. Microbial profiles in saliva from children with and without caries in mixed dentition. Oral Dis. 2012, 18, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Wade, W. Unculturable bacteria—The uncharacterized organisms that cause oral infections. J. R. Soc. Med. 2002, 95, 81–83. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zeng, X.; Ning, K.; Liu, K.L.; Lo, C.C.; Wang, W.; Chen, J.; Wang, D.; Huang, R.; Chang, X.; et al. Saliva microbiomes distinguish caries-active from healthy human populations. ISME J. 2012, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Hao, W.; Zhou, Q.; Wang, W.; Xia, Z.; Liu, C.; Chen, X.; Qin, M.; Chen, F. Plaque bacterial microbiome diversity in children younger than 30 months with or without caries prior to eruption of second primary molars. PLoS ONE 2014, 9, e89269. [Google Scholar] [CrossRef] [PubMed]
- Paster, B.J.; Boches, S.K.; Galvin, J.L.; Ericson, R.E.; Lau, C.N.; Levanos, V.A.; Sahasrabudhe, A.; Dewhirst, F.E. Bacterial diversity in human subgingival plaque. J. Bacteriol. 2001, 183, 3770–3783. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; Li, Y.; Zhou, C.; Wong, D.T.W.; Ho, C.M.; Qi, F.; Shi, W. Bacterial 16s rRNA/rDNA profiling in the liquid phase of human saliva. Open Dent. J. 2009, 3, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Gross, E.L.; Beall, C.J.; Kutsch, S.R.; Firestone, N.D.; Leys, E.J.; Griffen, A.L. Beyond streptococcus mutans: Dental caries onset linked to multiple species by 16s rRNA community analysis. PLoS ONE 2012, 7, e47722. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than blast. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Hamady, M.; Lozupone, C.; Knight, R. Fast unifrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and phylochip data. ISME J. 2010, 4, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Schellenberg, J.; Links, M.G.; Hill, J.E.; Dumonceaux, T.J.; Peters, G.A.; Tyler, S.; Ball, T.B.; Severini, A.; Plummer, F.A. Pyrosequencing of the chaperonin—60 universal target as a tool for determining microbial community composition. Appl. Environ. Microbiol. 2009, 75, 2889–2898. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Zhou, Y.; Ouyang, Y.; Lin, H. Dynamics of oral microbial community profiling during severe early childhood caries development monitored by PCR-DGGE. Arch. Oral Biol. 2013, 58, 1129–1138. [Google Scholar] [CrossRef] [PubMed]
- Consortium, H.M.P. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [Green Version]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16s rrna gene sequencing on the illumina miseq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Qin, M.; Chen, F.; Xia, B. Supragingival microbial profiles of permanent and deciduous teeth in children with mixed dentition. PLoS ONE 2016, 11, e0146938. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, R.; Lappin, D.F.; Dixon, P.M.; Buijs, M.J.; Zaura, E.; Crielaard, W.; O’Donnell, L.; Bennett, D.; Brandt, B.W.; Riggio, M.P. The microbiome associated with equine periodontitis and oral health. Vet. Res. 2016, 47, 49. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Kong, J.; Jia, P.; Wei, C.; Wang, Y.; Pan, Z.; Huang, W.; Li, L.; Chen, H.; Xiang, C. Analysis of oral microbiota in children with dental caries by PCR-DGGE and barcoded pyrosequencing. Microb. Ecol. 2010, 60, 677–690. [Google Scholar] [CrossRef] [PubMed]
- Zaura, E.; Keijser, B.J.; Huse, S.M.; Crielaard, W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009, 9, 259. [Google Scholar] [CrossRef] [PubMed]
- Milnes, A.R.; Bowden, G.H. The microflora associated with developing lesions of nursing caries. Caries Res. 1985, 19, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Loesche, W.J. Role of streptococcus mutans in human dental decay. Microbiol. Rev. 1986, 50, 353–380. [Google Scholar] [PubMed]
- Van Houte, J.; Jordan, H.V.; Laraway, R.; Kent, R.; Soparkar, P.M.; DePaola, P.F. Association of the microbial flora of dental plaque and saliva with human root-surface caries. J. Dent. Res. 1990, 69, 1463–1468. [Google Scholar] [CrossRef] [PubMed]
- Tanner, A.C.; Mathney, J.M.; Kent, R.L.; Chalmers, N.I.; Hughes, C.V.; Loo, C.Y.; Pradhan, N.; Kanasi, E.; Hwang, J.; Dahlan, M.A.; et al. Cultivable anaerobic microbiota of severe early childhood caries. J. Clin. Microbiol. 2011, 49, 1464–1474. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Jiang, N.; Wang, S.; Hu, X.; Jiao, K.; He, X.; Li, Z.; Wang, J. Exploration of human salivary microbiomes—Insights into the novel characteristics of microbial community structure in caries and caries-free subjects. PLoS ONE 2016, 11, e0147039. [Google Scholar] [CrossRef] [PubMed]
- Munson, M.A.; Banerjee, A.; Watson, T.F.; Wade, W.G. Molecular analysis of the microflora associated with dental caries. J. Clin. Microbiol. 2004, 42, 3023–3029. [Google Scholar] [CrossRef] [PubMed]
- Bizhang, M.; Ellerbrock, B.; Preza, D.; Raab, W.; Singh, P.; Beikler, T.; Henrich, B.; Zimmer, S. Detection of nine microorganisms from the initial carious root lesions using a TaqMan-based real-time PCR. Oral Dis. 2011, 17, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Suzuki, M.; Huang, Y.; Umeda, M.; Ishikawa, I.; Benno, Y. Prevotella shahii sp. nov. and Prevotella salivae sp. nov., isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 2004, 54, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Ramos, C.P.; Falsen, E.; Alvarez, N.; Akervall, E.; Sjoden, B.; Collins, M.D. Actinomyces graevenitzii sp. nov., isolated from human clinical specimens. Int. J. Syst. Bacteriol. 1997, 47, 885–888. [Google Scholar] [CrossRef] [PubMed]
- Belstrom, D.; Fiehn, N.E.; Nielsen, C.H.; Holmstrup, P.; Kirkby, N.; Klepac-Ceraj, V.; Paster, B.J.; Twetman, S. Altered bacterial profiles in saliva from adults with caries lesions: A case-cohort study. Caries Res. 2014, 48, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Hill, T.C.; Walsh, K.A.; Harris, J.A.; Moffett, B.F. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 2003, 43, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Hou, B. Analysis of microbial diversity of supra-gingival plaque in dental caries. Zhonghua Kou Qiang Yi Xue Za Zhi. 2014, 49, 742–747. [Google Scholar] [PubMed]
- World Health Organization (WHO). Oral Health Surveys: Basic Methods, 4th ed.; World Health Organization: Geneva, Switzerland, 1997. [Google Scholar]
- Quinque, D.; Kittler, R.; Kayser, M.; Stoneking, M.; Nasidze, I. Evaluation of saliva as a source of human DNA for population and association studies. Anal. Biochem. 2006, 353, 272–277. [Google Scholar] [CrossRef] [PubMed]

| Subject Code | dmft | Gender | Age | Reads after Trimming | OTUs |
|---|---|---|---|---|---|
| CA 1 | 2 | Male | 3 | 25,576 | 1974 |
| CA 2 | 2 | Male | 3 | 16,157 | 2452 |
| CA 3 | 2 | Female | 4 | 18,200 | 2306 |
| CA 4 | 2 | Female | 3 | 19,108 | 2304 |
| CA 5 | 10 | Male | 4 | 18,413 | 1919 |
| CA 6 | 16 | Female | 3 | 17,718 | 2123 |
| CA 7 | 2 | Male | 3 | 16,232 | 2591 |
| CA 8 | 2 | Male | 3 | 18,760 | 2445 |
| CA 9 | 6 | Female | 4 | 19,230 | 2201 |
| CA 10 | 7 | Male | 3 | 16,157 | 2770 |
| CA 11 | 2 | Female | 3 | 17,075 | 2254 |
| CA 12 | 2 | Male | 3 | 17,786 | 2403 |
| CA 13 | 2 | Male | 4 | 16,123 | 2306 |
| CA 14 | 2 | Female | 4 | 15,562 | 2310 |
| CA 15 | 2 | Male | 3 | 17,903 | 2535 |
| CA 16 | 2 | Female | 3 | 17,142 | 2225 |
| CA 17 | 2 | Male | 3 | 19,079 | 2190 |
| CA 18 | 2 | Female | 4 | 17,764 | 2398 |
| CA 19 | 2 | Female | 3 | 18,463 | 2476 |
| CA 20 | 10 | Female | 3 | 16,591 | 2134 |
| CF 1 | 0 | Male | 3 | 27,683 | 1979 |
| CF 2 | 0 | Male | 3 | 29,705 | 2306 |
| CF 3 | 0 | Male | 4 | 18,148 | 1559 |
| CF 4 | 0 | Female | 3 | 19,484 | 2025 |
| CF 5 | 0 | Female | 3 | 19,253 | 2370 |
| CF 6 | 0 | Female | 3 | 17,744 | 2212 |
| CF 7 | 0 | Male | 4 | 20,320 | 2639 |
| CF 8 | 0 | Male | 3 | 19,140 | 2376 |
| CF 9 | 0 | Female | 4 | 17,343 | 2127 |
| CF 10 | 0 | Female | 3 | 18,482 | 2133 |
| CF 11 | 0 | Male | 3 | 19,452 | 1796 |
| CF 12 | 0 | Male | 4 | 18,148 | 2130 |
| CF 13 | 0 | Male | 4 | 18,538 | 2259 |
| CF 14 | 0 | Male | 4 | 18,751 | 2416 |
| CF 15 | 0 | Female | 3 | 18,602 | 2122 |
| CF 16 | 0 | Female | 3 | 17,385 | 2406 |
| CF 17 | 0 | Female | 3 | 18,353 | 2334 |
| CF 18 | 0 | Female | 3 | 18,456 | 2702 |
| CF 19 | 0 | Male | 3 | 16,986 | 2610 |
| CF 20 | 0 | Female | 4 | 16,582 | 2684 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, S.; Gao, X.; Jin, L.; Lo, E.C.M. Salivary Microbiome Diversity in Caries-Free and Caries-Affected Children. Int. J. Mol. Sci. 2016, 17, 1978. https://doi.org/10.3390/ijms17121978
Jiang S, Gao X, Jin L, Lo ECM. Salivary Microbiome Diversity in Caries-Free and Caries-Affected Children. International Journal of Molecular Sciences. 2016; 17(12):1978. https://doi.org/10.3390/ijms17121978
Chicago/Turabian StyleJiang, Shan, Xiaoli Gao, Lijian Jin, and Edward C. M. Lo. 2016. "Salivary Microbiome Diversity in Caries-Free and Caries-Affected Children" International Journal of Molecular Sciences 17, no. 12: 1978. https://doi.org/10.3390/ijms17121978
APA StyleJiang, S., Gao, X., Jin, L., & Lo, E. C. M. (2016). Salivary Microbiome Diversity in Caries-Free and Caries-Affected Children. International Journal of Molecular Sciences, 17(12), 1978. https://doi.org/10.3390/ijms17121978







