IBTK Differently Modulates Gene Expression and RNA Splicing in HeLa and K562 Cells
Abstract
:1. Introduction
2. Results
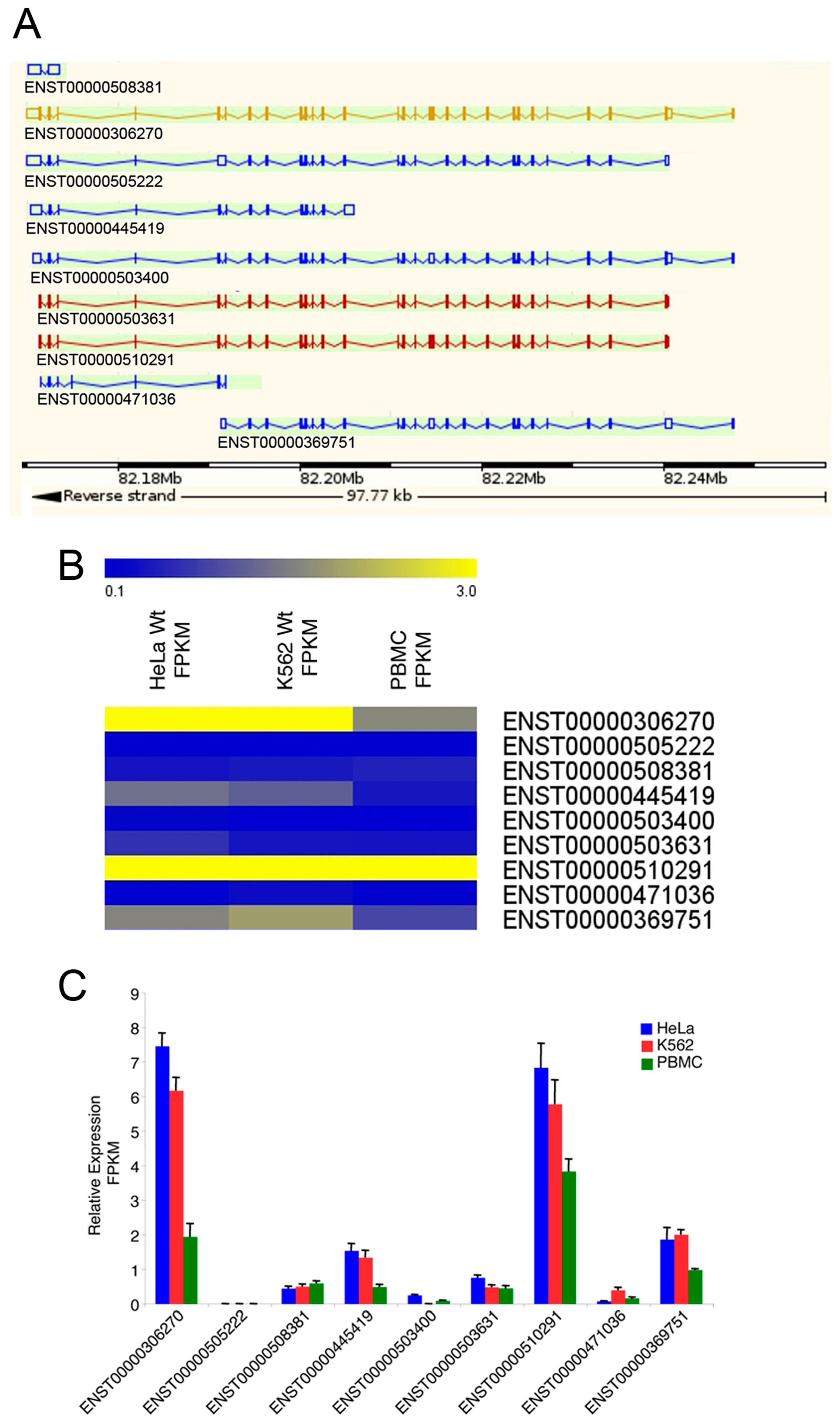
2.1. Expression Profile of the IBTK Gene in Different Cellular Contexts
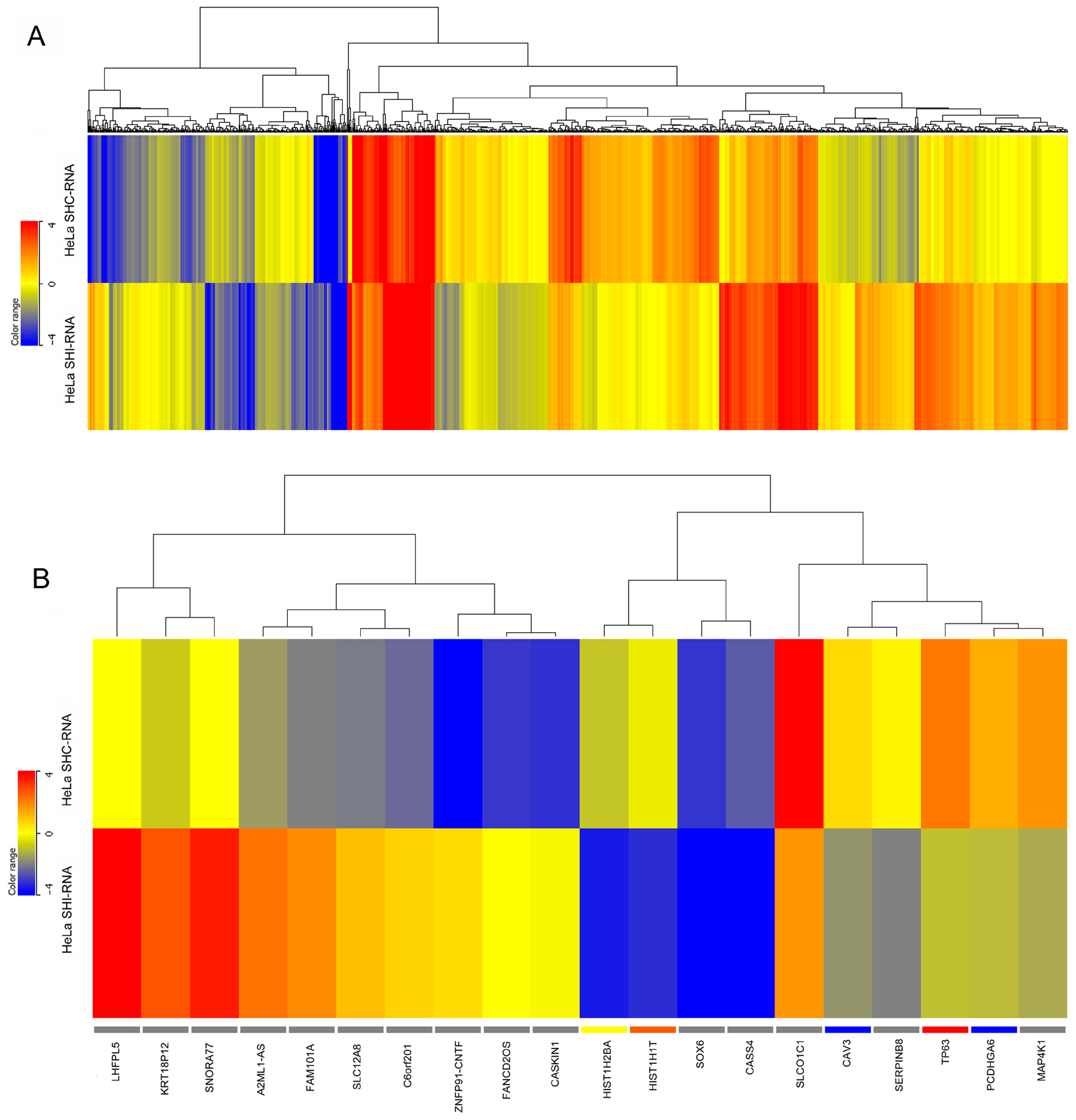
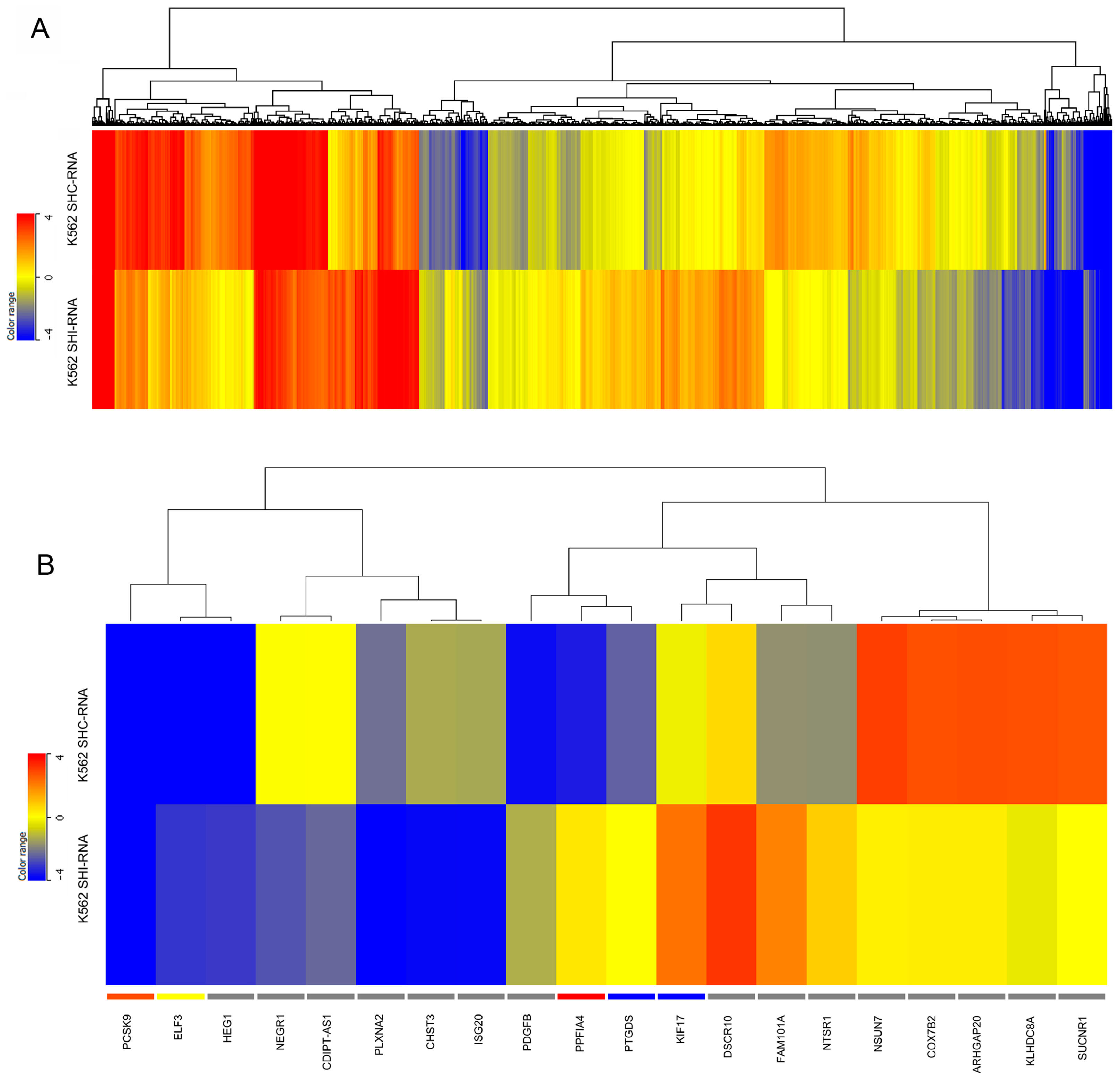
2.2. Differential Gene Expression in IBTK-Silenced HeLa and K562 Cells
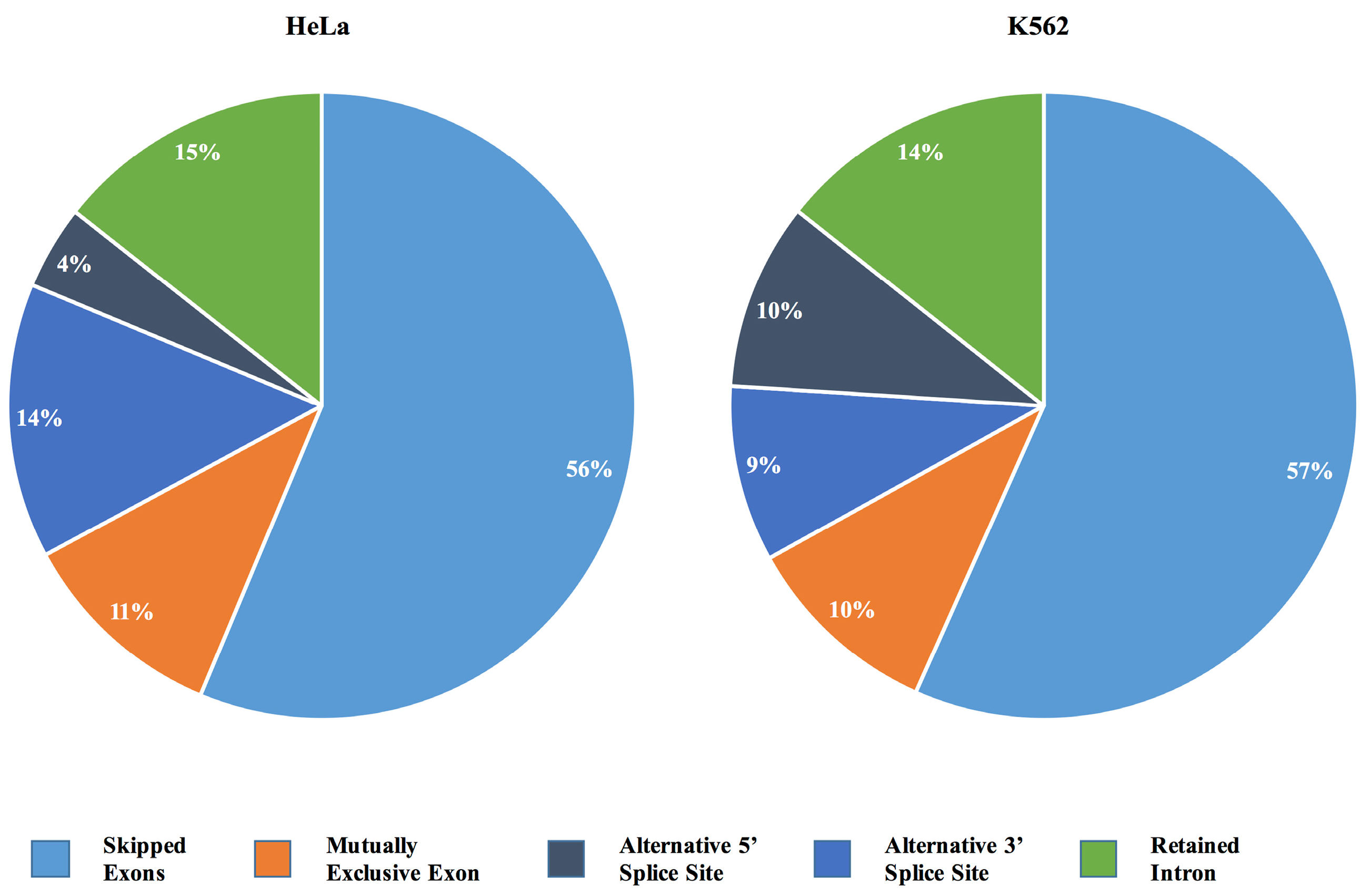
2.3. IBTK Affects Splicing Events in HeLa and K562 Cells
3. Discussion
4. Experimental Sections
4.1. Cells
4.2. Plasmids and Lentiviral Infections
4.3. Cells Extracts and Western Blotting
4.4. RNA Sequencing
4.5. Ethics Statement
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Spatuzza, C.; Schiavone, M.; di Salle, E.; Janda, E.; Sardiello, M.; Fiume, G.; Fierro, O.; Simonetta, M.; Argiriou, N.; Faraonio, R.; et al. Physical and functional characterization of the genetic locus of IBtk, an inhibitor of bruton’s tyrosine kinase: Evidence for three protein isoforms of IBtk. Nucleic Acids Res. 2008, 36, 4402–4416. [Google Scholar] [CrossRef]
- Janda, E.; Palmieri, C.; Pisano, A.; Pontoriero, M.; Iaccino, E.; Falcone, C.; Fiume, G.; Gaspari, M.; Nevolo, M.; di Salle, E.; et al. Btk regulation in human and mouse B cells via protein kinase C phosphorylation of IBtkγ. Blood 2011, 117, 6520–6531. [Google Scholar] [CrossRef]
- Fiume, G.; Rossi, A.; di Salle, E.; Spatuzza, C.; Mallardo, M.; Scala, G.; Quinto, I. Computational analysis and in vivo validation of a microRNA encoded by the IBTK gene, a regulator of B-lymphocytes differentiation and survival. Comput. Biol. Chem. 2009, 33, 434–439. [Google Scholar] [CrossRef]
- Liu, W.; Quinto, I.; Chen, X.; Palmieri, C.; Rabin, R.L.; Schwartz, O.M.; Nelson, D.L.; Scala, G. Direct inhibition of bruton’s tyrosine kinase by IBtk, a Btk-binding protein. Nat. Immunol. 2001, 2, 939–946. [Google Scholar] [CrossRef]
- Li, J.; Mahajan, A.; Tsai, M.D. Ankyrin repeat: A unique motif mediating protein-protein interactions. Biochemistry 2006, 45, 15168–15178. [Google Scholar] [CrossRef]
- Mosavi, L.K.; Cammett, T.J.; Desrosiers, D.C.; Peng, Z.Y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004, 13, 1435–1448. [Google Scholar] [CrossRef]
- Hadjebi, O.; Casas-Terradellas, E.; Garcia-Gonzalo, F.R.; Rosa, J.L. The RCC1 superfamily: From genes, to function, to disease. Biochim. Biophys. Acta 2008, 1783, 1467–1479. [Google Scholar] [CrossRef]
- Stogios, P.J.; Downs, G.S.; Jauhal, J.J.; Nandra, S.K.; Prive, G.G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005, 6, R82. [Google Scholar] [CrossRef]
- Chen, H.Y.; Chen, R.H. Cullin 3 ubiquitin ligases in cancer biology: Functions and therapeutic implications. Front. Oncol. 2016, 6, 113. [Google Scholar] [CrossRef]
- Pisano, A.; Ceglia, S.; Palmieri, C.; Vecchio, E.; Fiume, G.; de Laurentiis, A.; Mimmi, S.; Falcone, C.; Iaccino, E.; Scialdone, A.; et al. CRL3IBTK regulates the tumor suppressor Pdcd4 through ubiquitylation coupled to proteasomal degradation. J. Biol. Chem. 2015, 290, 13958–13971. [Google Scholar] [CrossRef]
- Lankat-Buttgereit, B.; Goke, R. The tumour suppressor Pdcd4: Recent advances in the elucidation of function and regulation. Biol. Cell 2009, 101, 309–317. [Google Scholar] [CrossRef]
- Ahmad, K.F.; Melnick, A.; Lax, S.; Bouchard, D.; Liu, J.; Kiang, C.L.; Mayer, S.; Takahashi, S.; Licht, J.D.; Prive, G.G. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol. Cell 2003, 12, 1551–1564. [Google Scholar] [CrossRef]
- Melnick, A.; Ahmad, K.F.; Arai, S.; Polinger, A.; Ball, H.; Borden, K.L.; Carlile, G.W.; Prive, G.G.; Licht, J.D. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol. Cell. Biol. 2000, 20, 6550–6567. [Google Scholar] [CrossRef]
- Ziegelbauer, J.; Shan, B.; Yager, D.; Larabell, C.; Hoffmann, B.; Tjian, R. Transcription factor MIZ-1 is regulated via microtubule association. Mol. Cell 2001, 8, 339–349. [Google Scholar] [CrossRef]
- Luo, J.; Emanuele, M.J.; Li, D.; Creighton, C.J.; Schlabach, M.R.; Westbrook, T.F.; Wong, K.K.; Elledge, S.J. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the ras oncogene. Cell 2009, 137, 835–848. [Google Scholar] [CrossRef]
- Kim, T.H.; Shin, S.W.; Park, J.S.; Park, C.S. Genome wide identification and expression profile in epithelial cells exposed to TiO2 particles. Environ. Toxicol. 2015, 30, 293–300. [Google Scholar] [CrossRef]
- Baird, T.D.; Palam, L.R.; Fusakio, M.E.; Willy, J.A.; Davis, C.M.; McClintick, J.N.; Anthony, T.G.; Wek, R.C. Selective mRNA translation during eIF2 phosphorylation induces expression of IBTKα. Mol. Biol. Cell 2014, 25, 1686–1697. [Google Scholar] [CrossRef]
- Broseus, J.; Chen, G.; Hergalant, S.; Ramstein, G.; Mounier, N.; Gueant, J.L.; Feugier, P.; Gisselbrecht, C.; Thieblemont, C.; Houlgatte, R. Relapsed diffuse large B-cell lymphoma present different genomic profiles between early and late relapses. Oncotarget 2016. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szczesniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef]
- Shen, S.; Park, J.W.; Huang, J.; Dittmar, K.A.; Lu, Z.X.; Zhou, Q.; Carstens, R.P.; Xing, Y. Mats: A bayesian framework for flexible detection of differential alternative splicing from RNA-seq data. Nucleic Acids Res. 2012, 40, e61. [Google Scholar] [CrossRef]
- Park, J.W.; Tokheim, C.; Shen, S.; Xing, Y. Identifying differential alternative splicing events from rna sequencing data using RNAseq-mats. Methods Mol. Biol. 2013, 1038, 171–179. [Google Scholar]
- Park, J.W.; Jung, S.; Rouchka, E.C.; Tseng, Y.T.; Xing, Y. rMAPS: RNA map analysis and plotting server for alternative exon regulation. Nucleic Acids Res. 2016, 44, W333–W338. [Google Scholar] [CrossRef]
- Hwang, S.K.; Baker, A.R.; Young, M.R.; Colburn, N.H. Tumor suppressor PDCD4 inhibits NF-κB-dependent transcription in human glioblastoma cells by direct interaction with p65. Carcinogenesis 2014, 35, 1469–1480. [Google Scholar] [CrossRef]
- Dago, D.N.; Scafoglio, C.; Rinaldi, A.; Memoli, D.; Giurato, G.; Nassa, G.; Ravo, M.; Rizzo, F.; Tarallo, R.; Weisz, A. Estrogen receptor beta impacts hormone-induced alternative mRNA splicing in breast cancer cells. BMC Genom. 2015, 16, 367. [Google Scholar] [CrossRef]
- Fiume, G.; Rossi, A.; de Laurentiis, A.; Falcone, C.; Pisano, A.; Vecchio, E.; Pontoriero, M.; Scala, I.; Scialdone, A.; Masci, F.F.; et al. Eukaryotic initiation factor 4H is under transcriptional control of p65/NF-κB. PLoS ONE 2013, 8, e66087. [Google Scholar] [CrossRef]
- Schiavone, M.; Fiume, G.; Caivano, A.; de Laurentiis, A.; Falcone, C.; Masci, F.F.; Iaccino, E.; Mimmi, S.; Palmieri, C.; Pisano, A.; et al. Design and characterization of a peptide mimotope of the HIV-1 gp120 bridging sheet. Int. J. Mol. Sci. 2012, 13, 5674–5699. [Google Scholar] [CrossRef]
- Tuccillo, F.M.; Palmieri, C.; Fiume, G.; de Laurentiis, A.; Schiavone, M.; Falcone, C.; Iaccino, E.; Galandrini, R.; Capuano, C.; Santoni, A.; et al. Cancer-associated CD43 glycoforms as target of immunotherapy. Mol. Cancer Ther. 2014, 13, 752–762. [Google Scholar] [CrossRef]
- Vitagliano, L.; Fiume, G.; Scognamiglio, P.L.; Doti, N.; Cannavo, R.; Puca, A.; Pedone, C.; Scala, G.; Quinto, I.; Marasco, D. Structural and functional insights into IκB-α/HIV-1 Tat interaction. Biochimie 2011, 93, 1592–1600. [Google Scholar] [CrossRef]
- De Laurentiis, A.; Gaspari, M.; Palmieri, C.; Falcone, C.; Iaccino, E.; Fiume, G.; Massa, O.; Masullo, M.; Tuccillo, F.M.; Roveda, L.; et al. Mass spectrometry-based identification of the tumor antigen UN1 as the transmembrane CD43 sialoglycoprotein. Mol. Cell. Proteom. MCP 2011, 10, M111.007898. [Google Scholar] [CrossRef]
- Fiume, G.; Scialdone, A.; Albano, F.; Rossi, A.; Tuccillo, F.M.; Rea, D.; Palmieri, C.; Caiazzo, E.; Cicala, C.; Bellevicine, C.; et al. Impairment of T cell development and acute inflammatory response in HIV-1 tat transgenic mice. Sci. Rep. 2015, 5, 13864. [Google Scholar] [CrossRef] [Green Version]
- Catalano, S.; Campana, A.; Giordano, C.; Gyorffy, B.; Tarallo, R.; Rinaldi, A.; Bruno, G.; Ferraro, A.; Romeo, F.; Lanzino, M.; et al. Expression and function of phosphodiesterase type 5 in human breast cancer cell lines and tissues: Implications for targeted therapy. Clin. Cancer Res. 2016, 22, 2271–2282. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiume, G.; Scialdone, A.; Rizzo, F.; De Filippo, M.R.; Laudanna, C.; Albano, F.; Golino, G.; Vecchio, E.; Pontoriero, M.; Mimmi, S.; et al. IBTK Differently Modulates Gene Expression and RNA Splicing in HeLa and K562 Cells. Int. J. Mol. Sci. 2016, 17, 1848. https://doi.org/10.3390/ijms17111848
Fiume G, Scialdone A, Rizzo F, De Filippo MR, Laudanna C, Albano F, Golino G, Vecchio E, Pontoriero M, Mimmi S, et al. IBTK Differently Modulates Gene Expression and RNA Splicing in HeLa and K562 Cells. International Journal of Molecular Sciences. 2016; 17(11):1848. https://doi.org/10.3390/ijms17111848
Chicago/Turabian StyleFiume, Giuseppe, Annarita Scialdone, Francesca Rizzo, Maria Rosaria De Filippo, Carmelo Laudanna, Francesco Albano, Gaetanina Golino, Eleonora Vecchio, Marilena Pontoriero, Selena Mimmi, and et al. 2016. "IBTK Differently Modulates Gene Expression and RNA Splicing in HeLa and K562 Cells" International Journal of Molecular Sciences 17, no. 11: 1848. https://doi.org/10.3390/ijms17111848
APA StyleFiume, G., Scialdone, A., Rizzo, F., De Filippo, M. R., Laudanna, C., Albano, F., Golino, G., Vecchio, E., Pontoriero, M., Mimmi, S., Ceglia, S., Pisano, A., Iaccino, E., Palmieri, C., Paduano, S., Viglietto, G., Weisz, A., Scala, G., & Quinto, I. (2016). IBTK Differently Modulates Gene Expression and RNA Splicing in HeLa and K562 Cells. International Journal of Molecular Sciences, 17(11), 1848. https://doi.org/10.3390/ijms17111848











