Association Analysis of Noncoding Variants in Neuroligins 3 and 4X Genes with Autism Spectrum Disorder in an Italian Cohort
Abstract
:1. Introduction
2. Results
2.1. Single-Locus Analysis
2.2. Haplotype Analysis (EUR Control Population)
2.3. Haplotype Analysis (ITA control Population)
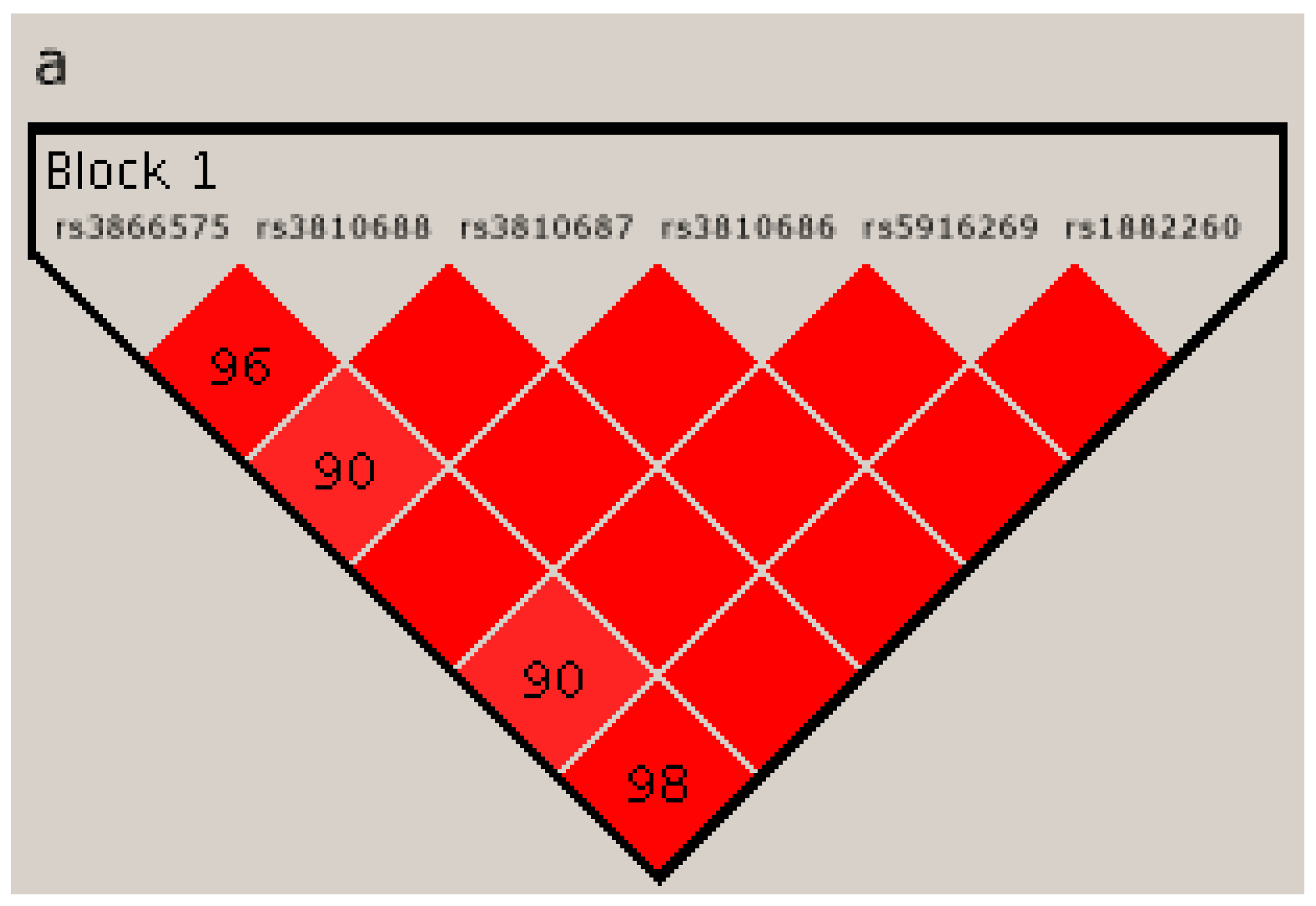
2.4. Linkage Disequilibrium Analysis
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. Selection of SNPs
4.3. SNPs Screening and Genotyping
4.4. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ASD | Autism spectrum disorder |
| EUR | European |
| FDR | False discovery rate |
| HRM | High resolution melting |
| IQ | Intelligence Quotient |
| ITA | Italian |
| MAF | Minor allele frequency |
| NLGN | Neuroligin |
| NSMR | Non-syndromic mental retardation |
| PDD-NOS | Pervasive developmental disorder-not otherwise specified |
| SNP | Single nucleotide polymorphism |
References
- Abrahams, B.S.; Geschwind, D.H. Advances in autism genetics: On the threshold of a new neurobiology. Nat. Rev. 2008, 9, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.P. Myers SM: Identification and evaluation of children with autism spectrum disorder. Pediatrics 2007, 120, 188–197. [Google Scholar] [CrossRef] [PubMed]
- London, E.; Etzel, R.A. The environment as an etiologic factor in autism: A new direction for research. Environ. Health Perspect. 2000, 108, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Tordjman, S.; Somogyi, E.; Coulon, N.; Kermarrec, S.; Cohen, D.; Bronsard, G.; Bonnot, O.; Weismann-Arcache, C.; Botbol, M.; Lauth, B.; et al. Gene × environment interactions in autism spectrum disorders: Role of epigenetic mechanisms. Front. Psychiatr. 2014, 5, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mezzelani, A.; Landini, M.; Facchiano, F.; Raggi, M.E.; Villa, L.; Molteni, M.; de Santis, B.; Brera, C.; Caroli, A.M.; Milanesi, L.; et al. Environment, dysbiosis, immunity and sex-specific susceptibility: An evidence-based translational hypothesis for regressive autism pathogenesis. Nutr. Neurosci. 2014, 1, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; He, X.; Yao, D.; Li, Z.; Zhao, Z. A sex specific association of common variants of neuroligin genes (NLGN3 and NLGN4X) with autism spectrum disorders in a Chinese Han cohort. Behav. Brain Funct. 2011, 7, 13–23. [Google Scholar] [CrossRef] [PubMed]
- David, B.; Scherer, S.W. Genetic architecture in autism spectrum disorder. Curr. Opin. Genet. Dev. 2012, 22, 229–237. [Google Scholar]
- Talkowsky, M.E.; Minikel, E.V.; Gusella, J.F. Autism spectrum disorder genetics: Diverse genes with diverse clinical outcomes. Harv. Rev. Psychiatr. 2014, 22, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Piton, A.; Gauthier, J.; Hamdam, F.F.; Lafreniere, R.G.; Yang, Y.; Henrion, E.; Laurent, S.; Noreau, A.; Thibodeau, P.; Karemera, L.; et al. Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder ad schizophrenia. Mol. Psychiatry 2011, 16, 867–880. [Google Scholar] [CrossRef] [PubMed]
- Vorstman, J.A.; Staal, W.G.; van Daalen, E.; van Engeland, H.; Hochstenbach, P.F.; Franke, L. Identification of novel autism candidate regions through the analysis of reported cytogenetic abnormalities associated with autism. Mol. Psychiatr. 2006, 11, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Betancur, C. Etiological heterogeneity in autism spectrum disorders: More than 100 genetic and genomic disorders and still counting. Brain Res. 2011, 1380, 42–77. [Google Scholar] [CrossRef] [PubMed]
- Jamain, S.; Quach, H.; Betancur, C.; Råstam, M.; Colineaux, C.; Gillberg, I.C.; Soderstrom, H.; Giros, B.; Leboyer, M.; Gillberg, C.; et al. Paris autism research international sibpair study: Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 2003, 34, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Laumonnier, F.; Bonnet-Brilhault, F.; Gomot, M.; Blanc, R.; David, A.; Moizard, M.P.; Raynaud, M.; Ronce, N.; Lemonnier, E.; Calvas, P.; et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am. J. Hum. Genet. 2004, 74, 552–557. [Google Scholar] [CrossRef] [PubMed]
- Shibayama, A.; Cook, E.H.; Feng, J.; Glanzmann, C.; Yan, J.; Craddock, N.; Jones, I.R.; Goldman, D.; Heston, L.L.; Sommer, S.S. MECP2 structural and 3′ UTR variants in schizophrenia, autism and other psychiatric diseases; a possible association with autism. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2004, 128, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Oliveira, G.; Coutinho, A.; Yang, C.; Feng, J.; Katz, C.; Sram, J.; Bockholt, A.; Jones, I.R.; Craddock, N.; et al. Analysis of the neuroligin 3 and 4 genes in autism and other neuropsychiatric patients. Mol. Psychiatr. 2005, 10, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Chih, B.; Engelman, H.; Scheiffele, P. Control of excitatory and inhibitory synapse formation by neuroligins. Science 2005, 307, 1324–1328. [Google Scholar] [CrossRef] [PubMed]
- Lisè, M.F.; El-Husseini, A. The neuroligin and neurexin families: From structure to function at the synapse. Cell. Mol. Life Sci. 2006, 63, 1833–1849. [Google Scholar] [CrossRef] [PubMed]
- Varoqueaux, F.; Aramuni, G.; Rawson, R.L.; Mohrmann, R.; Missler, M.; Gottmann, K.; Zhang, W.; Südhof, T.C.; Brose, N. Neuroligins determine synapse maturation and function. Neuron 2006, 51, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Pettem, K.L.; Yokomaku, D.; Takahashi, H.; Ge, Y.; Craig, A.M. Interaction between autism-linked MDGAs and neuroligins suppresses inhibitory synapse development. J. Cell Biol. 2013, 200, 321–336. [Google Scholar] [CrossRef] [PubMed]
- Südhof, T.C. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 2008, 455, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Zoghbi, H.Y.; Bear, M.F. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef] [PubMed]
- Chih, B.; Afridi, S.K.; Clark, L.; Scheiffele, P. Disorder associated mutations lead to functional inactivation of neuroligins. Hum. Mol. Genet. 2004, 13, 1471–1477. [Google Scholar] [CrossRef] [PubMed]
- Comoletti, D.; De Jaco, A.; Jennings, L.L.; Flynn, R.E.; Gaietta, G.; Tsigelny, I.; Ellisman, M.H.; Taylor, P. The Arg451Cys-neuroligin 3 mutation associated with autism reveals a defect in protein processing. J. Neurosci. 2004, 24, 4889–4893. [Google Scholar] [CrossRef] [PubMed]
- Rothwell, P.E.; Fuccillo, M.V.; Maxeiner, S.; Hayton, S.J.; Gokce, O.; Lim, B.K.; Fowler, S.C.; Malenka, R.C.; Südhof, T.C. Autism-associated neuroligin 3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell 2014, 158, 198–212. [Google Scholar] [CrossRef] [PubMed]
- Tabuchi, K.; Blundell, J.; Etherton, M.R.; Hammer, R.E.; Liu, X.; Powell, C.M.; Südhof, T.C. A neuroligin 3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science 2007, 318, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Jamain, S.K.; Hammerschmidt, R.K.; Granon, S.; Boretius, S.; Varoqueaux, F.; Ramanantsoa, N.; Gallego, J.; Ronnenberg, A.; Winter, D.; Frahm, J.; et al. Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proc. Natl. Acad. Sci. USA 2008, 105, 1710–1715. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Milunsky, J.M.; Newton, S.; Ko, J.; Zhao, G.; Maher, T.A.; Tager-Flusberg, H.; Bolliger, M.F.; Carter, A.S.; Boucard, A.A.; et al. A neuroligin 4 missense mutation associated with autism impairs neuroligin 4 folding and endoplasmic reticulum export. J. Neurosci. 2009, 29, 10843–10854. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Xiong, Z.; Zhang, L.; Liu, Y.; Lu, L.; Peng, Y.; Guo, H.; Zhao, J.; Xia, K.; Hu, Z. Variations analysis of NLGN3 and NLGN4X gene in Chinese autism patients. Mol. Biol. Rep. 2014, 41, 4133–4140. [Google Scholar]
- Talebizadeh, Z.; Lam, D.Y.; Theodoro, M.F.; Bittel, D.C.; Lushington, G.H.; Butler, M.G. Novel splice isoforms for NLGN3 and NLGN4 with possible implication in autism. J. Med. Genet. 2006, 43, e21. [Google Scholar] [CrossRef] [PubMed]
- Lawson-Yuen, A.; Saldivar, J.S.; Sommer, S.; Picker, J. Familial deletion within NLGN4 associated with autism and Tourette syndrome. Eur. J. Hum. Genet. 2008, 16, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Ylisaukko-oja, T.; Rehnström, K.; Auranen, M.; Vanhala, R.; Alen, R.; Kempas, E.; Ellonen, P.; Turunen, J.A.; Makkonen, I.; Riikonen, R.; et al. Analysis of four neuroligin genes as candidates for autism. Eur. J. Hum. Genet. 2005, 13, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Daoud, H.; Bonnet-Brilhault, F.; Vesdrine, S.; Demattei, M.V.; Vourc’h, P.; Bayou, N.; Andres, C.R.; Barthélémy, C.; Laumonnier, F.; Briault, S. Autism and nonsyndromic mental retardation associated with a de novo mutation in the NLGN4X gene promoter causing an increased expression level. Soc. Biol. Psychiatry 2009, 66, 906–910. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, K.M.; Ramachandran, D.; Patel, V.C.; Shetty, A.C.; Cutler, D.J.; Zwick, M.E. Identification of rare X-linked neuroligin variants by massively parallel sequencing in males with autism spectrum disorder. Mol. Autism 2012, 3, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Yanagi, K.; Kaname, T.; Wakui, K.; Hashimoto, O.; Fukushima, Y.; Naritomi, K. Identification of four novel synonymous substitutions in neuroligin 3 and neuroligin 4X in Japanase patients with autistic spectrum disorder. Autism Res. Treat. 2012, 2012, 724072–724077. [Google Scholar] [PubMed]
- Volaki, K.; Pampanos, A.; Kitsiou-Tzeli, S.; Vrettou, C.; Oikonomakis, V.; Sofocleous, C.; Kanavakis, E. Mutation screening in the Greek population and evaluation of NLGN3 and NLGN4X genes causal factors for autism. Psychiatr. Genet. 2013, 23, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Xing, L.; Zhang, K.; Gao, X.; Zheng, Z.; Huang, S.; Guo, Y.; Zhang, F. Positive association of neuroigin-4 gene with non specific mental retardation in the Qinba mountains region of China. Psychiatr. Genet. 2009, 19, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Gao, X.; Qi, H.; Zheng, Z.; Zhang, F. Gender differencs in cognitive ability associated with genetic variants of NLGN4. Neuropsychiatry 2010, 62, 221–228. [Google Scholar]
- Liu, Y.; Du, Y.; Liu, W.; Yang, C.; Liu, Y.; Wang, H.; Gong, X. Lack of association between NLGN3, NLGN4, SHANK2 and SHANK3 gene variants and autism spectrum disorder in a Chinese population. PLoS ONE 2013, 8, e56639. [Google Scholar] [CrossRef] [PubMed]
- Blasi, F.; Bacchelli, E.; Pesaresi, G.; Carone, S.; Bailey, A.J.; Maestrini, E. International molecular genetic study of autism consortium (IMGAC): Absence of coding mutations in the X-linked genes neuroligin 3 and neuroligin 4 in individuals with autism from the IMGSAC collection. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006, 141, 220–221. [Google Scholar] [CrossRef] [PubMed]
- Wermenter, A.K.; Kamp-Becker, I.; Strauch, K.; Schulte-Korne, G.; Remschmidt, H. No evidence for involvement of genetic variants in the X-linked neuroligin genes NLGN3 and NLGN4X in probands with autism spectrum disorder on high functioning level. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008, 147, 535–537. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, J.; Bonnel, A.; St-Onge, J.; Karamera, L.; Laurent, S.; Mottron, L.; Fombonne, E.; Joober, R.; Rouleau, G.A. NLGN3/NLGN4 gene mutations are not responsible for autism in the Quebec population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005, 132, 74–75. [Google Scholar] [CrossRef] [PubMed]
- Altshuler, D.; Durbin, R.M.; Abecasis, G.R.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Collins, F.S.; De La Vega, F.M.; Donnelly, P.; Egholm, M.; et al. A map of human genome variation from population-scale sequencing. Nature 2010, 467, 1061–1073. [Google Scholar] [Green Version]
- Salvi, E.; Kutalik, Z.; Glorioso, N.; Benaglio, P.; Frau, F.; Kuznetsova, T.; Arima, H.; Hoggart, C.; Tichet, J.; Nikitin, Y.P.; et al. Genome wide association study using a high-density single nucleotide polymorphism array and case-control design identifies a novel essential hypertension susceptibility locus in the promoter region of endothelial NO synthase. Hypertension 2012, 59, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Howie, B.; Fuchsberger, C.; Stephens, M.; Marchini, J.; Abecasis, G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012, 44, 955–959. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]

| Gene ID | SNP ID | Minor Allele | MAF Controls | MAF Case | p-Value | OR | 95% CI | p-Corr |
|---|---|---|---|---|---|---|---|---|
| NLGN3 | rs11795613 | G | 0.481 | 0.515 | 0.398 | 1.144 | 0.816–1.617 | 0.701 |
| rs4844285 | A | 0.4741 | 0.4895 | 0.701 | 1.064 | 0.755–1.496 | 0.701 | |
| rs4844286 | T | 0.505 | 0.481 | 0.5396 | 0.908 | 0.646–1.280 | 0.701 | |
| NLGN4X | rs6638575 | A | 0.266 | 0.301 | 0.303 | 1.193 | 0.856–1.663 | 0.404 |
| rs3810688 | T | 0.2176 | 0.288 | 0.0456 | 1.453 | 1.018–2.074 | 0.152 | |
| rs3810687 | T | 0.1086 | 0.1339 | 0.3369 | 1.269 | 0.805–2.000 | 0.404 | |
| rs3810686 | T | 0.4466 | 0.4561 | 0.817 | 1.039 | 0.768–1.407 | 0.817 | |
| rs5916269 | A | 0.1086 | 0.1339 | 0.3369 | 1.269 | 0.805–2.000 | 0.404 | |
| rs1882260 | C | 0.2655 | 0.3347 | 0.0505 | 1.392 | 1.005–1.928 | 0.152 |
| N | SNPs in Haplotypes | Control Frequency | Case Frequency | Haplotype | p-Value | Emp. p-Value | Emp. q-Value |
|---|---|---|---|---|---|---|---|
| 6 | rs6638575-rs3810688-rs3810687-rs3810686-rs5916269-rs1882260 | 0.0335 | 0.1799 | GTGCGT | 2.58 × 10−6 | <1 × 10−5 | <1 × 10−5 |
| 5 | rs6638575-rs3810688-rs3810687-rs3810686-rs5916269 | 0.0805 | 0.1799 | GTGCG | 7.60 × 10−6 | <1 × 10−5 | <1 × 10−5 |
| 5 | rs3810688-rs3810687-rs3810686-rs5916269-rs1882260 | 0.0335 | 0.1799 | TGCGT | 2.58 × 10−6 | <1 × 10−5 | <1 × 10−5 |
| 4 | rs6638575-rs3810688-rs3810687-rs3810686 | 0.0805 | 0.1799 | GTGC | 7.6 × 10−6 | <1 × 10−5 | 2 × 10−5 |
| 4 | rs3810688-rs3810687-rs3810686-rs5916269 | 0.0801 | 0.1793 | TGCG | 7.6 × 10−6 | <1 × 10−5 | 2 × 10−5 |
| 4 | rs3810687-rs3810686-rs5916269-rs1882260 | 0.0943 | 0.1793 | GCGT | 1.3 × 10−5 | <1 × 10−5 | 2 × 10−5 |
| 3 | rs6638575-rs3810688-rs3810687 | 0.3538 | 0.4463 | GTG | 8.16 × 10−6 | <1 × 10−5 | <1 × 10−5 |
| 3 | rs3810688-rs3810687-rs3810686 | 0.0701 | 0.1793 | TGC | 7.6 × 10−6 | <1 × 10−5 | <1 × 10−5 |
| 3 | rs3810687-rs3810686-rs5916269 | 0.4062 | 0.4445 | GCG | 8.16 × 10−4 | 6.99 × 10−4 | 3.2 × 10−3 |
| 3 | rs3810686-rs5916269-rs1882260 | 0.0943 | 0.1793 | CGT | 1.3 × 10−5 | <1 × 10−5 | 3 × 10−4 |
| 2 | rs6638575-rs3810688 | 0.2109 | 0.2854 | GT | 1.08 × 10−4 | 7 × 10−5 | 8.3 × 10−4 |
| 2 | rs3810688-rs3810687 | 0.0943 | 0.1793 | TG | 1.3 × 10−5 | 2 × 10−5 | 1.5 × 10−4 |
| 2 | rs3810687-rs3810686 | 0.4062 | 0.4445 | GC | 8.16 × 10−4 | 7.3 × 10−4 | 5 × 10−3 |
| 2 | rs3810686-rs5916269 | 0.4062 | 0.4445 | CG | 8.16 × 10−4 | 7.3 × 10−4 | 5 × 10−3 |
| 2 | rs5916269-rs1882260 | 0.4314 | 0.6259 | GT | 2.4 × 10−8 | <1 × 10−5 | <1 × 10−5 |
| N | SNPs in Haplotypes | Control Frequency | Case Frequency | Haplotype | p-Value | Empirical p-Value | Empirical q-Value |
|---|---|---|---|---|---|---|---|
| 6 | rs6638575-rs3810688-rs3810687-rs3810686-rs5916269-rs1882260 | 0.0678 | 0.1799 | GTGCGT | 6.33 × 10−3 | 6.79 × 10−3 | 2.73 × 10−3 |
| 5 | rs6638575-rs3810688-rs3810687-rs3810686-rs5916269 | 0.1349 | 0.1799 | GTGCG | 1.9 × 10−2 | 1.87 × 10−2 | 4.47 × 10−2 |
| 5 | rs3810688-rs3810687-rs3810686-rs5916269-rs1882260 | 0.0678 | 0.1799 | TGCGT | 6.33 × 10−3 | 5.39 × 10−3 | 2.78 × 10−2 |
| 4 | rs6638575-rs3810688-rs3810687-rs3810686 | 0.1189 | 0.1799 | GTGC | 1.66 × 10−2 | 1.62 × 10−2 | 4.41 × 10−2 |
| 3 | rs6638575-rs3810688-rs3810687 | 0.3782 | 0.4463 | GTG | 1.56 × 10−2 | 1.29 × 10−2 | 4.37 × 10−2 |
| 3 | rs3810688-rs3810687-rs3810686 | 0.0801 | 0.1793 | TGC | 1.17 × 10−2 | 1.26 × 10−2 | 4.78 × 10−2 |
| 3 | rs3810687-rs3810686-rs5916269 | 0.3360 | 0.4445 | GCG | 3.99 × 10−3 | 4.10 × 10−3 | 1.98 × 10−2 |
| 2 | rs3810687-rs3810686 | 0.3160 | 0.4445 | GC | 3.50 × 10−3 | 3.20 × 10−3 | 2.20 × 10−2 |
| 2 | rs3810686-rs5916269 | 0.3360 | 0.4445 | CG | 3.99 × 10−3 | 3.70 × 10−3 | 2.53 × 10−2 |
| Patients: 202; Age: 2–12; Males: 165; Females: 37 | ||
|---|---|---|
| Patients from North Italy: n = 157 | Males: 131, 83.44% | Young autism: 93 (71%) |
| PDD-NOS: 34 (26%) | ||
| Asperger’s Syndrome: 4 (3%) | ||
| Females: 26, 16.56% | Young autism: 18 (69.2%) | |
| PDD-NOS: 7 (27%) | ||
| Asperger’s Syndrome: 1 (3.8%) | ||
| Patients from South Italy: n = 45 | Males: 34, 75.55% | Young autism: 24 (70.6%) |
| PDD-NOS: 9 (26.5%) | ||
| Asperger’s Syndrome: 1 (2.9%) | ||
| Females: 11, 24.45% | Young autism: 8 (72.7%) | |
| PDD-NOS: 3 (27.3%) | ||
| Asperger’s Syndrome: 0 | ||
| IQ | Hyperactivity | Language and Communication | |
|---|---|---|---|
| >70 n = 71 (35.1%) | Level 1: IQ > 101; n = 15 (7.4%) | Level 0: n = 136; (67.3%) | Level 1: n = 7 (3.5%) |
| Level 2: 100 > IQ> 70; n = 56 (27.7%) | Level 2: n = 48 (23.7%) | ||
| ≤69 n = 131 (64.6%) | Level 3: 69 > IQ > 50; n = 79 (39.1%) | Level 1: n = 53 (26.2%) | Level 3: n = 65 (32.2%) |
| Level 4: 49 > IQ > 35; n = 28 (13.9%) | Level 4: n = 64 (31.7%) | ||
| Level 5: 34 > IQ> 20; n = 17 (8.4%) | Level 2: n = 13 (6.5%) | Level 5: n = 3 (1.5%) | |
| Level 6: IQ < 19; n = 7 (3.5%) | 15 patients not evaluated (7.4%) | ||
| Genes | SNPs | Alleles | MAF | Position in Gene | References |
|---|---|---|---|---|---|
| NLGN3 | rs11795613 | (A/G) | G: 0.49 | Intron 1 | Yu et al., Behav. Brain Funct. 2011, 7, 13 [6] |
| rs4844285 | (A/G) | A: 0.48 | Intron 2 | ||
| rs4844286 | (T/G) | T: 0.49 | Intron 2 | ||
| NLGN4X | rs6638575 | (A/G) | A: 0.28 | Intron 5 | Qi et al., Psychiatr. Genet. 2009, 19, 1 [36] |
| rs3810686 | (T/C) | T: 0.44 | 3′ UTR | ||
| rs1882260 | (C/T) | C: 0.27 | 3′ UTR | ||
| rs3810687 | (T/G) | T: 0.11 | 3′ UTR | – | |
| rs3810688 | (T/C) | T: 0.29 | 3′ UTR | ||
| rs5916269 | (A/G) | A: 0.11 | 3′ UTR |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Landini, M.; Merelli, I.; Raggi, M.E.; Galluccio, N.; Ciceri, F.; Bonfanti, A.; Camposeo, S.; Massagli, A.; Villa, L.; Salvi, E.; et al. Association Analysis of Noncoding Variants in Neuroligins 3 and 4X Genes with Autism Spectrum Disorder in an Italian Cohort. Int. J. Mol. Sci. 2016, 17, 1765. https://doi.org/10.3390/ijms17101765
Landini M, Merelli I, Raggi ME, Galluccio N, Ciceri F, Bonfanti A, Camposeo S, Massagli A, Villa L, Salvi E, et al. Association Analysis of Noncoding Variants in Neuroligins 3 and 4X Genes with Autism Spectrum Disorder in an Italian Cohort. International Journal of Molecular Sciences. 2016; 17(10):1765. https://doi.org/10.3390/ijms17101765
Chicago/Turabian StyleLandini, Martina, Ivan Merelli, M. Elisabetta Raggi, Nadia Galluccio, Francesca Ciceri, Arianna Bonfanti, Serena Camposeo, Angelo Massagli, Laura Villa, Erika Salvi, and et al. 2016. "Association Analysis of Noncoding Variants in Neuroligins 3 and 4X Genes with Autism Spectrum Disorder in an Italian Cohort" International Journal of Molecular Sciences 17, no. 10: 1765. https://doi.org/10.3390/ijms17101765







