Viral Metagenomics on Blood-Feeding Arthropods as a Tool for Human Disease Surveillance
Abstract
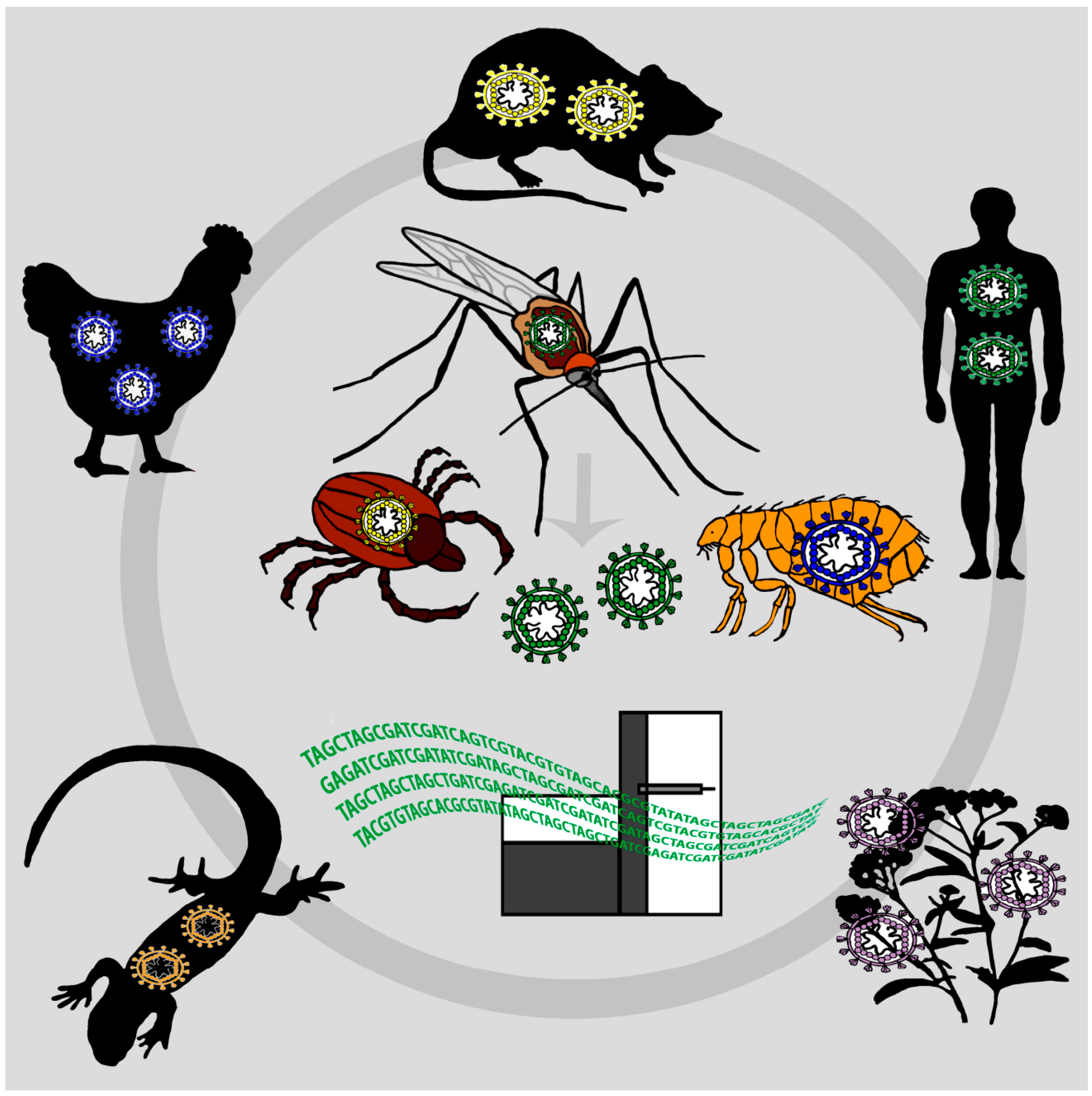
:1. Surveillance of Emerging Viruses
2. Mosquitoes as “Flying Syringes” for Virus Surveillance
2.1. Feasibility Studies
2.2. Other Hematophagous Animals for VEM
2.2.1. Ticks
2.2.2. Midges
2.2.3. Carrion Flies
2.2.4. Bed Bugs
2.2.5. Fleas
2.2.6. Bat Flies
2.2.7. Leeches (phylum Annelida)
2.2.8. Lice
3. Combining VEM with iDNA
4. Mosquito Sampling: Practical Considerations
5. Discussion
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Raj, V.S.; Osterhaus, A.D.; Fouchier, R.A.; Haagmans, B.L. Mers: Emergence of a novel human coronavirus. Curr. Opin. Virol. 2014, 5, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Li, D.X. Severe fever with thrombocytopenia syndrome: A newly discovered emerging infectious disease. Clin. Microbiol. Infect. 2015, 21, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Carroll, M.W.; Matthews, D.A.; Hiscox, J.A.; Elmore, M.J.; Pollakis, G.; Rambaut, A.; Hewson, R.; Garcia-Dorival, I.; Bore, J.A.; Koundouno, R.; et al. Temporal and spatial analysis of the 2014–2015 Ebola virus outbreak in West Africa. Nature 2015, 524, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Grard, G.; Fair, J.N.; Lee, D.; Slikas, E.; Steffen, I.; Muyembe, J.J.; Sittler, T.; Veeraraghavan, N.; Ruby, J.G.; Wang, C.; et al. A novel rhabdovirus associated with acute hemorrhagic fever in Central Africa. PLoS Pathog. 2012, 8, e1002924. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef] [PubMed]
- EcoHealth Alliance. EcoHealth Alliance Pandemic Prevention: PREDICT. Available online: http://www.ecohealthalliance.org/program/predict (accessed on 4 May 2016).
- Temmam, S.; Davoust, B.; Berenger, J.M.; Raoult, D.; Desnues, C. Viral metagenomics on animals as a tool for the detection of zoonoses prior to human infection? Int. J. Mol. Sci. 2014, 15, 10377–10397. [Google Scholar] [CrossRef] [PubMed]
- Molaei, G.; Andreadis, T.G.; Armstrong, P.M.; Anderson, J.F.; Vossbrinck, C.R. Host feeding patterns of Culex mosquitoes and West Nile virus transmission, Northeastern United States. Emerg. Infect. Dis. 2006, 12, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Grubaugh, N.D.; Sharma, S.; Krajacich, B.J.; Fakoli, L.S., III; Bolay, F.K.; Diclaro, J.W., II; Johnson, W.E.; Ebel, G.D.; Foy, B.D.; Brackney, D.E. Xenosurveillance: A novel mosquito-based approach for examining the human-pathogen landscape. PLoS Negl. Trop. Dis. 2015, 9, e0003628. [Google Scholar] [CrossRef] [PubMed]
- Delwart, E.L. Viral metagenomics. Rev. Med. Virol. 2007, 17, 115–131. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.F.; Willner, D.L.; Lim, Y.W.; Schmieder, R.; Chau, B.; Nilsson, C.; Anthony, S.; Ruan, Y.; Rohwer, F.; Breitbart, M. Broad surveys of DNA viral diversity obtained through viral metagenomics of mosquitoes. PLoS ONE 2011, 6, e20579. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.F.; Duffy, S.; Polston, J.E.; Bixby, E.; Vallad, G.E.; Breitbart, M. Exploring the diversity of plant DNA viruses and their satellites using vector-enabled metagenomics on whiteflies. PLoS ONE 2011, 6, e19050. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Liu, Y.; Hu, X.; Xiong, J.; Zhang, B.; Yuan, Z. A metagenomic survey of viral abundance and diversity in mosquitoes from Hubei province. PLoS ONE 2015, 10, e0129845. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.A.; Liu, R.M.; Bennett, S.N. RNA shotgun metagenomic sequencing of Northern California (USA) mosquitoes uncovers viruses, bacteria, and fungi. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Coffey, L.L.; Page, B.L.; Greninger, A.L.; Herring, B.L.; Russell, R.C.; Doggett, S.L.; Haniotis, J.; Wang, C.; Deng, X.; Delwart, E.L. Enhanced arbovirus surveillance with deep sequencing: Identification of novel rhabdoviruses and bunyaviruses in Australian mosquitoes. Virology 2014, 448, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Bishop-Lilly, K.A.; Turell, M.J.; Willner, K.M.; Butani, A.; Nolan, N.M.; Lentz, S.M.; Akmal, A.; Mateczun, A.; Brahmbhatt, T.N.; Sozhamannan, S.; et al. Arbovirus detection in insect vectors by rapid, high-throughput pyrosequencing. PLoS Negl. Trop. Dis. 2010, 4, e878. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Huang, Y.; Gong, Z.; Zhuang, L.; Li, C.; Yang, H.; Tong, Y.; Liu, W.; Cao, W. Discovery of DNA viruses in wild-caught mosquitoes using small RNA high throughput sequencing. PLoS ONE 2011, 6, e24758. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.A.; Thongsripong, P.; Green, A.; Kittayapong, P.; Wilcox, B.A.; Schroth, G.P.; Kapan, D.D.; Bennett, S.N. Metagenomic shotgun sequencing of a bunyavirus in wild-caught Aedes aegypti from Thailand informs the evolutionary and genomic history of the Phleboviruses. Virology 2014, 464–465, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Shi, M.; Tian, J.H.; Lin, X.D.; Kang, Y.J.; Chen, L.J.; Qin, X.C.; Xu, J.; Holmes, E.C.; Zhang, Y.Z. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Bolling, B.G.; Weaver, S.C.; Tesh, R.B.; Vasilakis, N. Insect-specific virus discovery: Significance for the arbovirus community. Viruses 2015, 7, 4911–4928. [Google Scholar] [CrossRef] [PubMed]
- Manzin, A.; Martina, B.E.; Gould, E.A.; Bagnarelli, P.; Sambri, V. Human arthropod-borne viral infections. BioMed Res. Int. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Gould, E.A.; Solomon, T. Pathogenic flaviviruses. Lancet 2008, 371, 500–509. [Google Scholar] [CrossRef]
- Zacks, M.A.; Paessler, S. Encephalitic alphaviruses. Vet. Microbiol. 2010, 140, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Guo, L.; Zhang, L.; Wei, Y.; Huang, L.; Wu, H.; Liu, C. Three new emerging subgroups of torque teno sus viruses (TTSuVs) and co-infection of TTSuVswith porcine circovirus type 2 in China. Virol. J. 2013, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Barbazan, P.; Thitithanyanont, A.; Misse, D.; Dubot, A.; Bosc, P.; Luangsri, N.; Gonzalez, J.P.; Kittayapong, P. Detection of H5N1 avian influenza virus from mosquitoes collected in an infected poultry farm in Thailand. Vector Borne Zoonotic Dis. 2008, 8, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Fields, B.N.; Knipe, D.M.; Howley, P.M. Fields Virology; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013. [Google Scholar]
- Burd, E.M. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 2003, 16, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kremsdorf, D.; Favre, M.; Jablonska, S.; Obalek, S.; Rueda, L.A.; Lutzner, M.A.; Blanchet-Bardon, C.; van Voorst Vader, P.C.; Orth, G. Molecular cloning and characterization of the genomes of nine newly recognized human papillomavirus types associated with epidermodysplasia verruciformis. J. Virol. 1984, 52, 1013–1018. [Google Scholar] [PubMed]
- Antonsson, A.; Forslund, O.; Ekberg, H.; Sterner, G.; Hansson, B.G. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. J. Virol. 2000, 74, 11636–11641. [Google Scholar] [CrossRef] [PubMed]
- Biagini, P.; Gallian, P.; Cantaloube, J.F.; Attoui, H.; de Micco, P.; de Lamballerie, X. Distribution and genetic analysis of TTV and TTMV major phylogenetic groups in French blood donors. J. Med. Virol. 2006, 78, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Biagini, P.; Uch, R.; Belhouchet, M.; Attoui, H.; Cantaloube, J.F.; Brisbarre, N.; de Micco, P. Circular genomes related to anelloviruses identified in human and animal samples by using a combined rolling-circle amplification/sequence-independent single primer amplification approach. J. Gen. Virol. 2007, 88, 2696–2701. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.F.; Suedmeyer, W.K.; Wheeler, E.; Gulland, F.; Breitbart, M. Novel anellovirus discovered from a mortality event of captive California sea lions. J. Gen. Virol. 2009, 90, 1256–1261. [Google Scholar] [CrossRef] [PubMed]
- Kading, R.C.; Biggerstaff, B.J.; Young, G.; Komar, N. Mosquitoes used to draw blood for arbovirus viremia determinations in small vertebrates. PLoS ONE 2014, 9, e99342. [Google Scholar] [CrossRef] [PubMed]
- Yuill, T.M. Mosquitoes for drawing blood from small reptiles. Trans. R. Soc. Trop. Med. Hyg. 1969, 63, 407–408. [Google Scholar] [CrossRef]
- Yajima, M.; Kanda, T.; Takada, K. Critical role of epstein-barr virus (EBV)-encoded RNA in efficient EBV-induced B-lymphocyte growth transformation. J. Virol. 2005, 79, 4298–4307. [Google Scholar] [CrossRef] [PubMed]
- Moormann, A.M.; Snider, C.J.; Chelimo, K. The company malaria keeps: How co-infection with epstein-barr virus leads to endemic burkitt lymphoma. Curr. Opin. Infect. Dis. 2011, 24, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Garver, L.S.; Bingham, K.M.; Hang, J.; Jochim, R.C.; Davidson, S.A.; Richardson, J.H.; Jarman, R.G. Feasibility of using the mosquito blood meal for rapid and efficient human and animal virus surveillance and discovery. Am. J. Trop. Med. Hyg. 2015, 93, 1377–1382. [Google Scholar] [CrossRef] [PubMed]
- Hubalek, Z.; Rudolf, I. Tick-borne viruses in Europe. Parasitol. Res. 2012, 111, 9–36. [Google Scholar] [CrossRef] [PubMed]
- Havlikova, S.; Lickova, M.; Klempa, B. Non-viraemic transmission of tick-borne viruses. Acta Virol. 2013, 57, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Bente, D.A.; Forrester, N.L.; Watts, D.M.; McAuley, A.J.; Whitehouse, C.A.; Bray, M. Crimean-congo hemorrhagic fever: History, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antivir. Res. 2013, 100, 159–189. [Google Scholar] [CrossRef] [PubMed]
- Tokarz, R.; Williams, S.H.; Sameroff, S.; Sanchez Leon, M.; Jain, K.; Lipkin, W.I. Virome analysis of amblyomma americanum, dermacentor variabilis, and ixodes scapularis ticks reveals novel highly divergent vertebrate and invertebrate viruses. J. Virol. 2014, 88, 11480–11492. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Hu, C.; Zhang, D.; Tang, S.; Zhang, Z.; Kou, Z.; Fan, Z.; Bente, D.; Zeng, C.; Li, T. Metagenomic profile of the viral communities in Rhipicephalus spp. Ticks from Yunnan, China. PLoS ONE 2015, 10, e0121609. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, J.M.; Ng, T.F.; Suzuki, Y.; Tsujimoto, H.; Deng, X.; Delwart, E.; Rasgon, J.L. Bunyaviruses are common in male and female Ixodes scapularis ticks in central Pennsylvania. PeerJ 2016, 4, e2324. [Google Scholar] [CrossRef] [PubMed]
- Moutailler, S.; Popovici, I.; Devillers, E.; Vayssier-Taussat, M.; Eloit, M. Diversity of viruses in ixodes ricinus, and characterization of a neurotropic strain of eyach virus. New Microbes New Infect. 2016, 11, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Obenchain, F.D.; Galun, R. (Eds.) Physiology of Ticks. Current Themes in Tropical Science; Pergamon Press: Oxford, UK, 1983; Volume 1.
- Gariepy, T.D.; Lindsay, R.; Ogden, N.; Gregory, T.R. Identifying the last supper: Utility of the DNA barcode library for bloodmeal identification in ticks. Mol. Ecol. Resour. 2012, 12, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Wodecka, B.; Rymaszewska, A.; Skotarczak, B. Host and pathogen DNA identification in blood meals of nymphal ixodes ricinus ticks from forest parks and rural forests of Poland. Exp. Appl. Acarol. 2013, 62, 543–555. [Google Scholar] [CrossRef] [PubMed]
- Ginsberg, H.S.; Ewing, C.P. Comparison of flagging, walking, trapping, and collecting from hosts as sampling methods for northern deer ticks, Ixodes dammini, and lone-star ticks, Amblyomma Americanum (Acari: Ixodidae). Exp. Appl. Acarol. 1989, 7, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, S.; Groschup, M.H.; Garros, C.; Felippe-Bauer, M.L.; Purse, B.V. Culicoides biting midges, arboviruses and public health in Europe. Antivir. Res. 2013, 100, 102–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Temmam, S.; Monteil-Bouchard, S.; Robert, C.; Baudoin, J.P.; Sambou, M.; Aubadie-Ladrix, M.; Labas, N.; Raoult, D.; Mediannikov, O.; Desnues, C. Characterization of viral communities of biting midges and identification of novel thogotovirus species and rhabdovirus genus. Viruses 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Beer, M.; Conraths, F.J.; van der Poel, W.H. “Schmallenberg virus”—A novel orthobunyavirus emerging in Europe. Epidemiol. Infect. 2013, 141, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Fall, M.; Diarra, M.; Fall, A.G.; Balenghien, T.; Seck, M.T.; Bouyer, J.; Garros, C.; Gimonneau, G.; Allene, X.; Mall, I.; et al. Culicoides (Diptera: Ceratopogonidae) midges, the vectors of african horse sickness virus—A host/vector contact study in the Niayes area of Senegal. Parasites Vectors 2015, 8. [Google Scholar] [CrossRef] [PubMed]
- Mellor, P.S.; Boorman, J.; Baylis, M. Culicoides biting midges: Their role as arbovirus vectors. Annu. Rev. Entomol. 2000, 45, 307–340. [Google Scholar] [CrossRef] [PubMed]
- Temmam, S.; Monteil-Bouchard, S.; Sambou, M.; Aubadie-Ladrix, M.; Azza, S.; Decloquement, P.; Khalil, J.Y.; Baudoin, J.P.; Jardot, P.; Robert, C.; et al. Faustovirus-like asfarvirus in hematophagous biting midges and their vertebrate hosts. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Sawabe, K.; Hoshino, K.; Isawa, H.; Sasaki, T.; Hayashi, T.; Tsuda, Y.; Kurahashi, H.; Tanabayashi, K.; Hotta, A.; Saito, T.; et al. Detection and isolation of highly pathogenic H5N1 avian influenza A viruses from blow flies collected in the vicinity of an infected poultry farm in Kyoto, Japan, 2004. Am. J. Trop. Med. Hyg. 2006, 75, 327–332. [Google Scholar] [PubMed]
- Chakrabarti, S.; King, D.J.; Afonso, C.; Swayne, D.; Cardona, C.J.; Kuney, D.R.; Gerry, A.C. Detection and isolation of exotic newcastle disease virus from field-collected flies. J. Med. Entomol. 2007, 44, 840–844. [Google Scholar] [CrossRef] [PubMed]
- Calibeo-Hayes, D.; Denning, S.S.; Stringham, S.M.; Guy, J.S.; Smith, L.G.; Watson, D.W. Mechanical transmission of Turkey coronavirus by domestic houseflies (Musca Domestica Linnaeaus). Avian Dis. 2003, 47, 149–153. [Google Scholar] [CrossRef]
- Tan, S.W.; Yap, K.L.; Lee, H.L. Mechanical transport of rotavirus by the legs and wings of musca domestica (Diptera: Muscidae). J. Med. Entomol. 1997, 34, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Schubert, G.; Stockhausen, M.; Hoffmann, C.; Merkel, K.; Vigilant, L.; Leendertz, F.H.; Calvignac-Spencer, S. Targeted detection of mammalian species using carrion fly-derived DNA. Mol. Ecol. Resour. 2015, 15, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Calvignac-Spencer, S.; Merkel, K.; Kutzner, N.; Kuhl, H.; Boesch, C.; Kappeler, P.M.; Metzger, S.; Schubert, G.; Leendertz, F.H. Carrion fly-derived DNA as a tool for comprehensive and cost-effective assessment of mammalian biodiversity. Mol. Ecol. 2013, 22, 915–924. [Google Scholar] [CrossRef] [PubMed]
- Goddard, J.; deShazo, R. Bed bugs (Cimex lectularius) and clinical consequences of their bites. JAMA 2009, 301, 1358–1366. [Google Scholar] [CrossRef] [PubMed]
- Delaunay, P.; Blanc, V.; del Giudice, P.; Levy-Bencheton, A.; Chosidow, O.; Marty, P.; Brouqui, P. Bedbugs and infectious diseases. Clin. Infect. Dis. 2011, 52, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Vail Mayans, M.; Hall, A.J.; Inskip, H.M.; Chotard, J.; Lindsay, S.W.; Alonso, P.L.; Coromina, E.; Mendy, M.; Whittle, H. Risk factors for transmission of hepatitis B virus to Gambian children. Lancet 1990, 336, 1107–1109. [Google Scholar] [CrossRef]
- Webb, P.A.; Happ, C.M.; Maupin, G.O.; Johnson, B.J.; Ou, C.Y.; Monath, T.P. Potential for insect transmission of HIV: Experimental exposure of cimex hemipterus and toxorhynchites amboinensis to human immunodeficiency virus. J. Infect. Dis. 1989, 160, 970–977. [Google Scholar] [CrossRef] [PubMed]
- Vobis, M.; D’Haese, J.; Mehlhorn, H.; Mencke, N. Evidence of horizontal transmission of feline leukemia virus by the cat flea (Ctenocephalides felis). Parasitol. Res. 2003, 91, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Mead-Briggs, A.R.; Vaughan, J.A. The differential transmissibility of myxoma virus strains of differing virulence grades by the rabbit flea spilopsyllus cuniculi (Dale). Epidemiol. Infect. 1975, 75, 237–247. [Google Scholar] [CrossRef]
- Dick, C.W.; Patterson, B.D. Bat flies: Obligate ectoparasites of bats. In Micromammals and Macroparasites: From Evolutionary Ecology to Management; Morand, S., Krasnov, B.R., Poulin, R., Eds.; Springer: Tokyo, Japan, 2006; pp. 179–194. [Google Scholar]
- Kohl, C.; Kurth, A. European bats as carriers of viruses with zoonotic potential. Viruses 2014, 6, 3110–3128. [Google Scholar] [CrossRef] [PubMed]
- Sawyer, R.T. Leech Biology and Behaviour: Feeding Biology, Ecology, and Systematics; Clarendon Press: Oxford, UK, 1986. [Google Scholar]
- Rocha, R.; Borda, E.; Andreone, F.; Rosa, G.M. First reports of leech parasitism in malagasy anurans. Comp. Parasitol. 2012, 79, 352–356. [Google Scholar] [CrossRef]
- Schnell, I.B.; Thomsen, P.F.; Wilkinson, N.; Rasmussen, M.; Jensen, L.R.; Willerslev, E.; Bertelsen, M.F.; Gilbert, M.T. Screening mammal biodiversity using DNA from leeches. Curr. Biol. 2012, 22, R262–R263. [Google Scholar] [CrossRef] [PubMed]
- Al-Khleif, A.; Roth, M.; Menge, C.; Heuser, J.; Baljer, G.; Herbst, W. Tenacity of mammalian viruses in the gut of leeches fed with porcine blood. J. Med. Microbiol. 2011, 60, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Shope, R.E. The leech as a potential virus reservoir. J. Exp. Med. 1957, 105, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Mann, K.H. Leeches (Hirudinea) Their Structure, Physiology, Ecology and Embryology with an Appendix on the Systematics of Marine Leeches; Mann, K.H., Knight-jones, W., Eds.; Pergamon Press: New York, NY, USA, 1962. [Google Scholar]
- Borda, E.; Oceguera-Figueroa, A.; Siddall, M.E. On the classification, evolution and biogeography of terrestrial haemadipsoid leeches (hirudinida: Arhynchobdellida: Hirudiniformes). Mol. Phylogenet. Evol. 2008, 46, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Temmam, S.; Monteil-Bouchard, S.; Robert, C.; Pascalis, H.; Michelle, C.; Jardot, P.; Charrel, R.; Raoult, D.; Desnues, C. Host-associated metagenomics: A guide to generating infectious RNA viromes. PLoS ONE 2015, 10, e0139810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calvignac-Spencer, S.; Leendertz, F.H.; Gilbert, M.T.; Schubert, G. An invertebrate stomach’s view on vertebrate ecology: Certain invertebrates could be used as “vertebrate samplers” and deliver DNA-based information on many aspects of vertebrate ecology. Bioessays 2013, 35, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Kent, R.J.; Norris, D.E. Identification of mammalian blood meals in mosquitoes by a multiplexed polymerase chain reaction targeting cytochrome B. Am. J. Trop. Med. Hyg. 2005, 73, 336–342. [Google Scholar] [PubMed]
- Kent, R.; Juliusson, L.; Weissmann, M.; Evans, S.; Komar, N. Seasonal blood-feeding behavior of culex tarsalis (Diptera: Culicidae) in weld county, Colorado, 2007. J. Med. Entomol. 2009, 46, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Bohmann, K.; Schnell, I.B.; Gilbert, M.T.P. When bugs reveal biodiversity. Mol. Ecol. 2013, 22, 909–911. [Google Scholar] [CrossRef] [PubMed]
- Lassen, S.B.; Nielsen, S.A.; Kristensen, M. Identity and diversity of blood meal hosts of biting midges (Diptera: Ceratopogonidae: Culicoides latreille) in Denmark. Parasites Vectors 2012, 5. [Google Scholar] [CrossRef] [PubMed]
- Schnell, I.B.; Sollmann, R.; Calvignac-Spencer, S.; Siddall, M.E.; Yu, D.W.; Wilting, A.; Gilbert, M.T. iDNA from terrestrial haematophagous leeches as a wildlife surveying and monitoring tool—Prospects, pitfalls and avenues to be developed. Front. Zool. 2015, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allan, B.F.; Goessling, L.S.; Storch, G.A.; Thach, R.E. Blood meal analysis to identify reservoir hosts for amblyomma americanum ticks. Emerg. Infect. Dis. 2010, 16, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Kent, R.J. Molecular methods for arthropod bloodmeal identification and applications to ecological and vector-borne disease studies. Mol. Ecol. Resour. 2009, 9, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Molaei, G.; Andreadis, T.G.; Armstrong, P.M.; Diuk-Wasser, M. Host-feeding patterns of potential mosquito vectors in connecticut, U.S.A.: Molecular analysis of bloodmeals from 23 species of Aedes, Anopheles, Culex, Coquillettidia, Psorophora, and Uranotaenia. J. Med. Entomol. 2008, 45, 1143–1151. [Google Scholar] [CrossRef]
- Pitzer, J.B.; Kaufman, P.E.; Tenbroeck, S.H.; Maruniak, J.E. Host blood meal identification by multiplex polymerase chain reaction for dispersal evidence of stable flies (Diptera:Muscidae) between livestock facilities. J. Med. Entomol. 2011, 48, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Thiemann, T.C.; Brault, A.C.; Ernest, H.B.; Reisen, W.K. Development of a high-throughput microsphere-based molecular assay to identify 15 common bloodmeal hosts of Culex mosquitoes. Mol. Ecol. Resour. 2012, 12, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Beier, J.C.; Perkins, P.V.; Wirtz, R.A.; Koros, J.; Diggs, D.; Gargan, T.P., II; Koech, D.K. Bloodmeal identification by direct enzyme-linked immunosorbent assay (ELISA), tested on Anopheles (Diptera: Culicidae) in Kenya. J. Med. Entomol. 1988, 25, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Logue, K.; Keven, J.B.; Cannon, M.V.; Reimer, L.; Siba, P.; Walker, E.D.; Zimmerman, P.A.; Serre, D. Unbiased characterization of Anopheles mosquito blood meals by targeted high-throughput sequencing. PLoS Negl. Trop. Dis. 2016, 10, e0004512. [Google Scholar] [CrossRef] [PubMed]
- Becker, N.; Petric, D.; Zgomba, M.; Boase, C.; Minoo, M.; Dahl, C.; Kaiser, A. Mosquitoes and Their Control; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Brown, H.E.; Paladini, M.; Cook, R.A.; Kline, D.; Barnard, D.; Fish, D. Effectiveness of mosquito traps in measuring species abundance and composition. J. Med. Entomol. 2008, 45, 517–521. [Google Scholar] [CrossRef]
- Vazquez-Prokopec, G.M.; Galvin, W.A.; Kelly, R.; Kitron, U. A new, cost-effective, battery-powered aspirator for adult mosquito collections. J. Med. Entomol. 2009, 46, 1256–1259. [Google Scholar] [CrossRef] [PubMed]
- Silver, J.B.; Service, M.W. Mosquito Ecology Field Sampling Methods; Springer: New York, NY, USA, 2008. [Google Scholar]
- Kent, R.J.; Thuma, P.E.; Mharakurwa, S.; Norris, D.E. Seasonality, blood feeding behavior, and transmission of plasmodium falciparum by Anopheles arabiensis after an extended drought in Southern Zambia. Am. J. Trop. Med. Hyg. 2007, 76, 267–274. [Google Scholar] [PubMed]
- Tirados, I.; Costantini, C.; Gibson, G.; Torr, S.J. Blood-feeding behaviour of the malarial mosquito Anopheles arabiensis: Implications for vector control. Med. Vet. Entomol. 2006, 20, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Nasci, R.S.; Savage, H.M.; White, D.J.; Miller, J.R.; Cropp, B.C.; Godsey, M.S.; Kerst, A.J.; Bennett, P.; Gottfried, K.; Lanciotti, R.S. West nile virus in overwintering Culex mosquitoes, New York City, 2000. Emerg. Infect. Dis. 2001, 7, 742–744. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, S.; Service, M.W. Studies on hibernating populations of the mosquito Culex pipiens L. In Southern and Northern England. J. Nat. Hist. 1983, 17, 849–857. [Google Scholar] [CrossRef]
- Takken, W.; Verhulst, N.O. Host preferences of blood-feeding mosquitoes. Annu. Rev. Entomol. 2013, 58, 433–453. [Google Scholar] [CrossRef] [PubMed]
- Kiszewski, A.; Mellinger, A.; Spielman, A.; Malaney, P.; Sachs, S.E.; Sachs, J. A global index representing the stability of malaria transmission. Am. J. Trop. Med. Hyg. 2004, 70, 486–498. [Google Scholar] [PubMed]
- Jensen, T.; Cockburn, A.F.; Kaiser, P.E.; Barnard, D.R. Human blood-feeding rates among sympatric sibling species of anopheles quadrimaculatus mosquitoes in Northern Florida. Am. J. Trop. Med. Hyg. 1996, 54, 523–525. [Google Scholar] [PubMed]
- Ramsdale, C.; Snow, K. Distribution of the genus anopheles in Europe. Eur. Mosq. Bull. 2000, 1–26. [Google Scholar]
- Scott, T.W.; Githeko, A.K.; Fleisher, A.; Harrington, L.C.; Yan, G. DNA profiling of human blood in anophelines from lowland and highland sites in Western Kenya. Am. J. Trop. Med. Hyg. 2006, 75, 231–237. [Google Scholar] [PubMed]
- McBride, C.S.; Baier, F.; Omondi, A.B.; Spitzer, S.A.; Lutomiah, J.; Sang, R.; Ignell, R.; Vosshall, L.B. Evolution of mosquito preference for humans linked to an odorant receptor. Nature 2014, 515, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.E.; Evans, B.R.; Zheng, W.; Obas, V.; Barrera-Martinez, L.; Egizi, A.; Zhao, H.; Caccone, A.; Powell, J.R. Human impacts have shaped historical and recent evolution in Aedes aegypti, the dengue and yellow fever mosquito. Evolution 2014, 68, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Walton, W.E.; Workman, P.D.; Tempelis, C.H. Dispersal, survivorship, and host selection of Culex erythrothorax (Diptera: Culicidae) associated with a constructed wetland in Southern California. J. Med. Entomol. 1999, 36, 30–40. [Google Scholar] [CrossRef] [PubMed]
- LaBeaud, A.D.; Sutherland, L.J.; Muiruri, S.; Muchiri, E.M.; Gray, L.R.; Zimmerman, P.A.; Hise, A.G.; King, C.H. Arbovirus prevalence in mosquitoes, Kenya. Emerg. Infect. Dis. 2011, 17, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Rohwer, F. Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques 2005, 39, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Kohl, C.; Brinkmann, A.; Dabrowski, P.W.; Radonic, A.; Nitsche, A.; Kurth, A. Protocol for metagenomic virus detection in clinical specimens. Emerg. Infect. Dis. 2015, 21, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Budhauliya, R.; Das, B.; Chatterjee, S.; Vanlalhmuaka; Veer, V. Next-generation sequencing in clinical virology: Discovery of new viruses. World J. Virol. 2015, 4, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Konishi, E. Size of blood meals of Aedes albopictus and Culex tritaeniorhynchus (Diptera: Culicidae) feeding on an unrestrained dog infected with Dirofilaria immitis (Spirurida: Filariidae). J. Med. Entomol. 1989, 26, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Ogunrinade, A. The measurement of blood meal size in Aedes aegypti (L.). Afr. J. Med. Med. Sci. 1980, 9, 69–71. [Google Scholar] [PubMed]
- Sinden, R.E. Plasmodium differentiation in the mosquito. Parassitologia 1999, 41, 139–148. [Google Scholar] [PubMed]
- Kraus, R.H.; van Hooft, P.; Waldenstrom, J.; Latorre-Margalef, N.; Ydenberg, R.C.; Prins, H.H. Avian influenza surveillance with FTA cards: Field methods, biosafety, and transportation issues solved. J. Vis. Exp. 2011. [Google Scholar] [CrossRef] [PubMed]
- Smit, P.W.; Elliott, I.; Peeling, R.W.; Mabey, D.; Newton, P.N. An overview of the clinical use of filter paper in the diagnosis of tropical diseases. Am. J. Trop. Med. Hyg. 2014, 90, 195–210. [Google Scholar] [CrossRef] [PubMed]
- Abdelwhab, E.M.; Luschow, D.; Harder, T.C.; Hafez, H.M. The use of FTA(r) filter papers for diagnosis of avian influenza virus. J. Virol. Methods 2011, 174, 120–122. [Google Scholar] [CrossRef] [PubMed]
- Brackney, D.E.; Isoe, J.; Black, W.C.; Zamora, J.; Foy, B.D.; Miesfeld, R.L.; Olson, K.E. Expression profiling and comparative analyses of seven midgut serine proteases from the yellow fever mosquito, Aedes aegypti. J. Insect Physiol. 2010, 56, 736–744. [Google Scholar] [CrossRef] [PubMed]
- Day, T.; Mideo, N.; Alizon, S. Why is HIV not vector-borne? Evol. Appl. 2008, 1, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Fredericks, D.N.; Relman, D.A. Sequence-based identification of microbial pathogens: A reconsideration of koch’s postulates. Clin. Microbiol. Rev. 1996, 9, 18–33. [Google Scholar]
- Mokili, J.L.; Rohwer, F.; Dutilh, B.E. Metagenomics and future perspectives in virus discovery. Curr. Opin. Virol. 2012, 2, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Shuda, M.; Chang, Y.; Moore, P.S. Clonal integration of a polyomavirus in human merkel cell carcinoma. Science 2008, 319, 1096–1100. [Google Scholar] [CrossRef] [PubMed]
- Morens, D.M.; Folkers, G.K.; Fauci, A.S. The challenge of emerging and re-emerging infectious diseases. Nature 2004, 430, 242–249. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brinkmann, A.; Nitsche, A.; Kohl, C. Viral Metagenomics on Blood-Feeding Arthropods as a Tool for Human Disease Surveillance. Int. J. Mol. Sci. 2016, 17, 1743. https://doi.org/10.3390/ijms17101743
Brinkmann A, Nitsche A, Kohl C. Viral Metagenomics on Blood-Feeding Arthropods as a Tool for Human Disease Surveillance. International Journal of Molecular Sciences. 2016; 17(10):1743. https://doi.org/10.3390/ijms17101743
Chicago/Turabian StyleBrinkmann, Annika, Andreas Nitsche, and Claudia Kohl. 2016. "Viral Metagenomics on Blood-Feeding Arthropods as a Tool for Human Disease Surveillance" International Journal of Molecular Sciences 17, no. 10: 1743. https://doi.org/10.3390/ijms17101743




