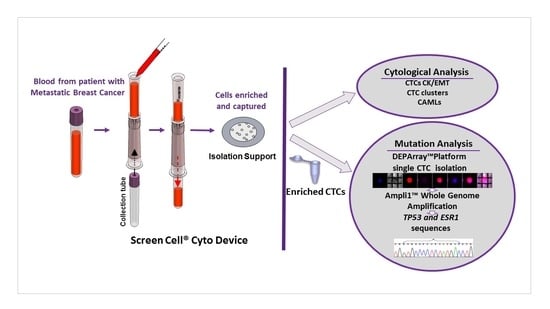
Detection and Characterization of Circulating Tumor Associated Cells in Metastatic Breast Cancer
Abstract
:1. Introduction
2. Results
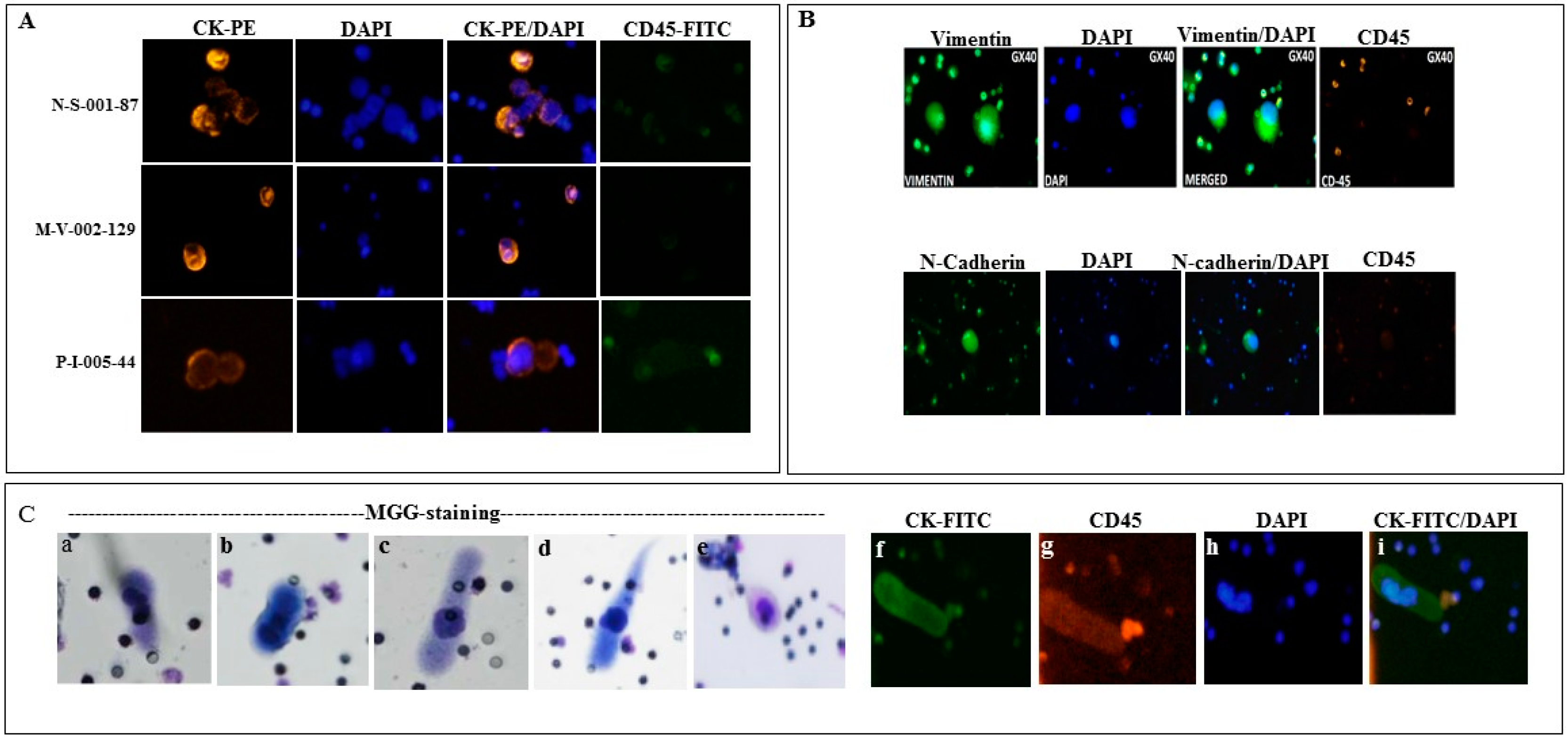
2.1. Detection of Circulating Tumor Associated Cells
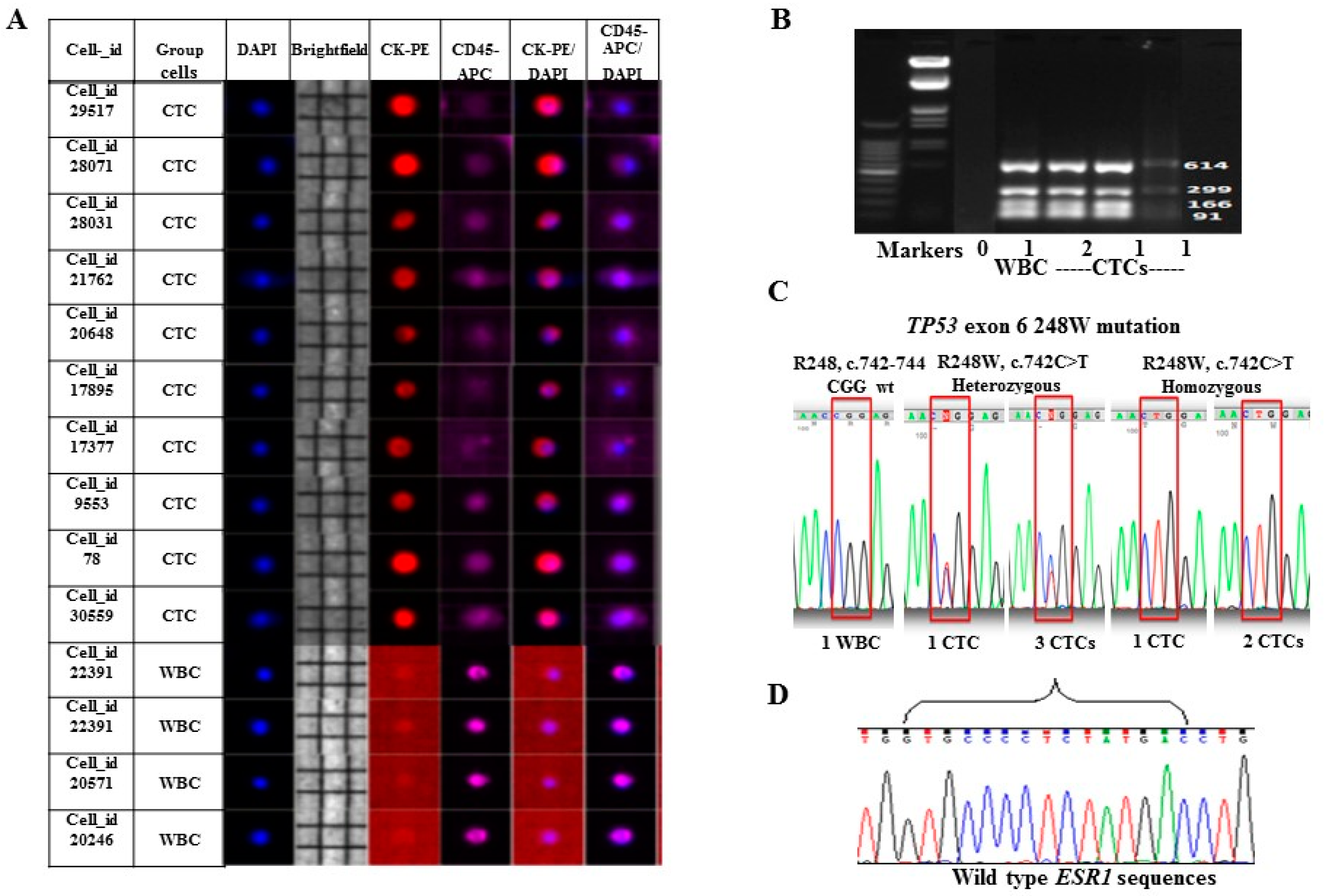
2.2. Enrichment, Single-Cell Sorting and Genome Amplification of Circulating Tumor Cells (CTCs)
2.3. Mutation Analysis of TP53 and ESR1 in Isolated CTCs
3. Discussion
4. Materials and Methods
4.1. Study Patients
4.2. Blood Sample Collection and Processing
4.3. Cytological Analysis
4.4. CTCs Enrichment and Single-Cell Sorting by DEPArray™ System
4.5. Ampli1™ Whole Genome Amplification and Quality Control Assays
4.6. TP53 and ESR1 Mutations in CTCs
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Angela, T.; Cristofanilli, M. Molecular characterization and targeted therapeutic approaches in breast cancer. Breast Cancer Res. 2015. [Google Scholar] [CrossRef]
- Krebs, M.G.; Metcalf, R.L.; Carter, L.; Brady, G.; Blackhall, F.H.; Dive, C. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat. Rev. Clin. Oncol. 2014, 11, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Lee, M.; Jeffrey, S.S. Circulating tumor cells and circulating tumor DNA: Challenges and opportunities on the path to clinical utility. Clin. Cancer Res. 2015, 21, 4786–4800. [Google Scholar] [CrossRef] [PubMed]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.; Uhr, J.W.; Terstappen, L.W. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [PubMed]
- Riethdorf, S.; Wikman, H.; Pantel, K. Review: Biological relevance of disseminated tumor cells in cancer patients. Int. J. Cancer 2008, 123, 1991–2006. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Bidard, F.C.; Peeters, D.J.; Fehm, T.; Nole, F.; Gisbert-Criado, R.; Mavroudis, D.; Grisanti, S.; Generali, D.; Garcia-Saenz, J.A.; Stebbing, J.; et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 2014, 15, 406–414. [Google Scholar] [CrossRef]
- Budd, G.T.; Cristofanilli, M.; Ellis, M.; Stopeck, A.; Borden, E.; Miller, M.C.; Matera, J.; Repollet, M.; Doyle, G.V.; Terstappen, L.W.; et al. Circulating tumor cells versus imaging-predicting overall survival in metastatic breast cancer. Clin. Cancer Res. 2006, 12, 6403–6409. [Google Scholar] [CrossRef] [PubMed]
- Hayes, D.F.; Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Miller, M.C.; Matera, J.; Allard, W.J.; Doyle, G.V.; Terstappen, L.W. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 2006, 12, 4218–4224. [Google Scholar] [CrossRef] [PubMed]
- Mego, M.; Mani, S.A.; Cristofanilli, M. Molecular mechanisms of metastasis in breast cancer-clinical applications. Nat. Rev. Clin. Oncol. 2010, 7, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Aktas, B.; Tewes, M.; Fehm, T.; Hauch, S.; Kimmig, R.; Kasimir-Bauer, S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009. [Google Scholar] [CrossRef] [PubMed]
- Papadaki, M.A.; Kallergi, G.; Zafeiriou, Z.; Manouras, L.; Theodoropoulos, P.A.; Mavroudis, D.; Georgoulias, V.; Agelaki, S. Co-expression of putative stemness and epithelial-to-mesenchymal transition markers on single circulating tumour cells from patients with early and metastatic breast cancer. BMC Cancer 2014. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.L.; Martin, S.S.; Alpaugh, R.K.; Charpentier, M.; Tsai, S.; Bergan, R.C.; Ogden, I.M.; Catalona, W.; Chumsri, S.; Tang, C.M.; et al. Circulating giant macrophages as a potential biomarker of solid tumors. Proc. Natl. Acad. Sci. USA. 2014, 111, 3514–3519. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 2014, 14, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Desitter, I.; Guerrouahen, B.S.; Benali-Furet, N.; Wechsler, J.; Jänne, P.A.; Kuang, Y.; Yanagita, M.; Wang, L.; Berkowitz, J.A.; Distel, R.J.; et al. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res. 2011, 31, 427–441. [Google Scholar] [PubMed]
- Chen, C.L.; Mahalingam, D.; Osmulski, P.; Jadhav, R.R.; Wang, C.M.; Leach, R.J.; Chang, T.C.; Weitman, S.D.; Kumar, A.P.; Sun, L.; et al. Single-cell analysis of circulating tumor cells identifies cumulative expression patterns of EMT-related genes in metastatic prostate cancer. Prostate 2013, 73, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Kulemann, B.; Pitman, M.B.; Liss, A.S.; Valsangkar, N.; Fernández-Del Castillo, C.; Lillemoe, K.D.; Hoeppner, J.; Mino-Kenudson, M.; Warshaw, A.L.; Thayer, S.P. Circulating tumor cells found in patients with localized and advanced pancreatic cancer. Pancreas 2015, 44, 547–550. [Google Scholar] [CrossRef] [PubMed]
- El-Heliebi, A.; Kroneis, T.; Zöhrer, E.; Haybaeck, J.; Fischereder, K.; Kampel-Kettner, K.; Zigeuner, R.; Pock, H.; Riedl, R.; Stauber, R.; et al. Are morphological criteria sufficient for the identification of circulating tumor cells in renal cancer? J. Transl. Med. 2013. [Google Scholar] [CrossRef] [PubMed]
- Coget, J.; Borrini, F.; Susman, S.; Sabourin, J.C. Colorectal carcinomas in 2013: The search for powerful prognostic markers is still on the go. Cancer Biomark. 2014, 14, 145–150. [Google Scholar] [PubMed]
- Mascalchi, M.; Falchini, M.; Maddau, C.; Salvianti, F.; Nistri, M.; Bertelli, E.; Sali, L.; Zuccherelli, S.; Vella, A.; Matucci, M.; et al. Prevalence and number of circulating tumour cells and microemboli at diagnosis of advanced NSCLC. J. Cancer Res. Clin. Oncol. 2016, 142, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Polzer, B.; Medoro, G.; Pasch, S.; Fontana, F.; Zorzino, L.; Pestka, A.; Andergassen, U.; Meier-Stiegen, F.; Czyz, Z.T.; Alberter, B.; et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol. Med. 2014, 30, 1371–1386. [Google Scholar] [CrossRef] [PubMed]
- Vishnoi, M.; Peddibhotla, S.; Yin, W.; Scamardo, A.T.; George, G.C.; Hong, D.S.; Marchetti, D. The isolation and characterization of CTC subsets related to breast cancer dormancy. Sci. Rep. 2015. [Google Scholar] [CrossRef] [PubMed]
- De Luca, F.; Rotunno, G.; Salvianti, F.; Galardi, F.; Pestrin, M.; Gabellini, S.; Simi, L.; Mancini, I.; Vannucchi, A.M.; Pazzagli, M.; et al. Mutational analysis of single circulating tumor cells by next generation sequencing in metastatic breast cancer. Oncotarget 2016. [Google Scholar] [CrossRef]
- Robinson, D.R.; Wu, Y.M.; Vats, P.; Su, F.; Lonigro, R.J.; Cao, X.; Kalyana-Sundaram, S.; Wang, R.; Ning, Y.; Hodges, L.; et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet. 2013, 45, 1446–1451. [Google Scholar] [CrossRef] [PubMed]
- Mu, Z.; Wang, C.; Ye, Z.; Austin, L.; Civan, J.; Hyslop, T.; Palazzo, J.P.; Jaslow, R.; Li, B.; Myers, R.E.; et al. Prospective assessment of the prognostic value of circulating tumor cells and their clusters in patients with advanced-stage breast cancer. Breast Cancer Res. Treat. 2015, 154, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Paoletti, C.; Li, Y.; Muniz, M.C.; Kidwell, K.M.; Aung, K.; Thomas, D.G.; Brown, M.E.; Abramson, V.G.; Irvin, W.J., Jr.; Lin, N.U.; et al. Significance of circulating tumor cells in metastatic triple-negative breast cancer patients within a randomized, phase II trial: TBCRC 019. Clin. Cancer Res. 2015, 21, 2771–2779. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sarkissyan, M.; Vadgama, J.V. Epithelial-mesenchymal transition and breast cancer. J. Clin. Med. 2016. [Google Scholar] [CrossRef] [PubMed]
- Kallergi, G.; Papadaki, M.A.; Politaki, E.; Mavroudis, D.; Georgoulias, V.; Agelaki, S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. 2011. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, A.J.; Marengo, M.S.; Oltean, S.; Kemeny, G.; Bitting, R.L.; Turnbull, J.D.; Herold, C.I.; Marcom, P.K.; George, D.J.; Garcia-Blanco, M.A. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol. Cancer Res. 2011, 9, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Kasimir-Bauer, S.; Hoffmann, O.; Wallwiener, D.; Kimmig, R.; Fehm, T. Expression of stem cell and epithelial-mesenchymal transition markers in primary breast cancer patients with circulating tumor cells. Breast Cancer Res. 2012. [Google Scholar] [CrossRef] [PubMed]
- Barrière, G.; Riouallon, A.; Renaudie, J.; Tartary, M.; Rigaud, M. Mesenchymal and stemness circulating tumor cells in early breast cancer diagnosis. BMC Cancer 2012. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, G.; Rath, B.; Klameth, L.; Hochmair, M.J. Small cell lung cancer: Recruitment of macrophages by circulating tumor cells. Oncoimmunology 2015. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.L.; Adams, D.K.; Alpaugh, R.K.; Cristofanilli, M.; Martin, S.S.; Chumsri, S.; Tang, C.M.; Marks, J.R. Circulating cancer-associated macrophage-like cells differentiate malignant breast cancer and benign breast conditions. Cancer Epidemiol. Biomark. Prev. 2016, 25, 1037–1042. [Google Scholar] [CrossRef] [PubMed]
- Smerage, J.B.; Barlow, W.E.; Hortobagyi, G.N.; Winer, E.P.; Leyland-Jones, B.; Srkalovic, G.; Tejwani, S.; Schott, A.F.; O’Rourke, M.A.; Lew, D.L.; et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J. Clin. Oncol. 2014, 32, 3483–3489. [Google Scholar] [CrossRef] [PubMed]

| Circulating Tumor Associated Cells | Total Patients (N) | Positive Patients (%) |
|---|---|---|
| CTCs ≥ 1 | 30 | 20 (66.7) |
| CTC clusters ≥ 2 CTCs | 30 | 8 (26.7) |
| EMT CTCs ≥ 1 | 28 | 13 (46.4) |
| CAMLs ≥ 1 | 20 | 15 (75.0) |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mu, Z.; Benali-Furet, N.; Uzan, G.; Znaty, A.; Ye, Z.; Paolillo, C.; Wang, C.; Austin, L.; Rossi, G.; Fortina, P.; et al. Detection and Characterization of Circulating Tumor Associated Cells in Metastatic Breast Cancer. Int. J. Mol. Sci. 2016, 17, 1665. https://doi.org/10.3390/ijms17101665
Mu Z, Benali-Furet N, Uzan G, Znaty A, Ye Z, Paolillo C, Wang C, Austin L, Rossi G, Fortina P, et al. Detection and Characterization of Circulating Tumor Associated Cells in Metastatic Breast Cancer. International Journal of Molecular Sciences. 2016; 17(10):1665. https://doi.org/10.3390/ijms17101665
Chicago/Turabian StyleMu, Zhaomei, Naoual Benali-Furet, Georges Uzan, Anaëlle Znaty, Zhong Ye, Carmela Paolillo, Chun Wang, Laura Austin, Giovanna Rossi, Paolo Fortina, and et al. 2016. "Detection and Characterization of Circulating Tumor Associated Cells in Metastatic Breast Cancer" International Journal of Molecular Sciences 17, no. 10: 1665. https://doi.org/10.3390/ijms17101665
APA StyleMu, Z., Benali-Furet, N., Uzan, G., Znaty, A., Ye, Z., Paolillo, C., Wang, C., Austin, L., Rossi, G., Fortina, P., Yang, H., & Cristofanilli, M. (2016). Detection and Characterization of Circulating Tumor Associated Cells in Metastatic Breast Cancer. International Journal of Molecular Sciences, 17(10), 1665. https://doi.org/10.3390/ijms17101665