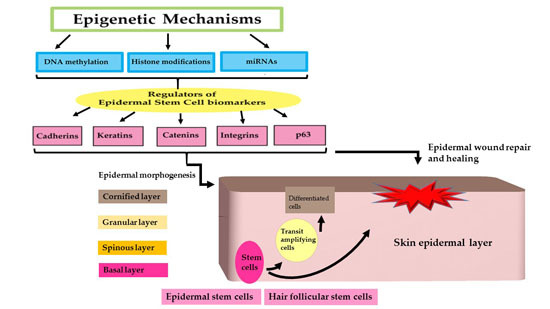
Epigenetic Regulation of Epidermal Stem Cell Biomarkers and Their Role in Wound Healing
Abstract
:1. Introduction
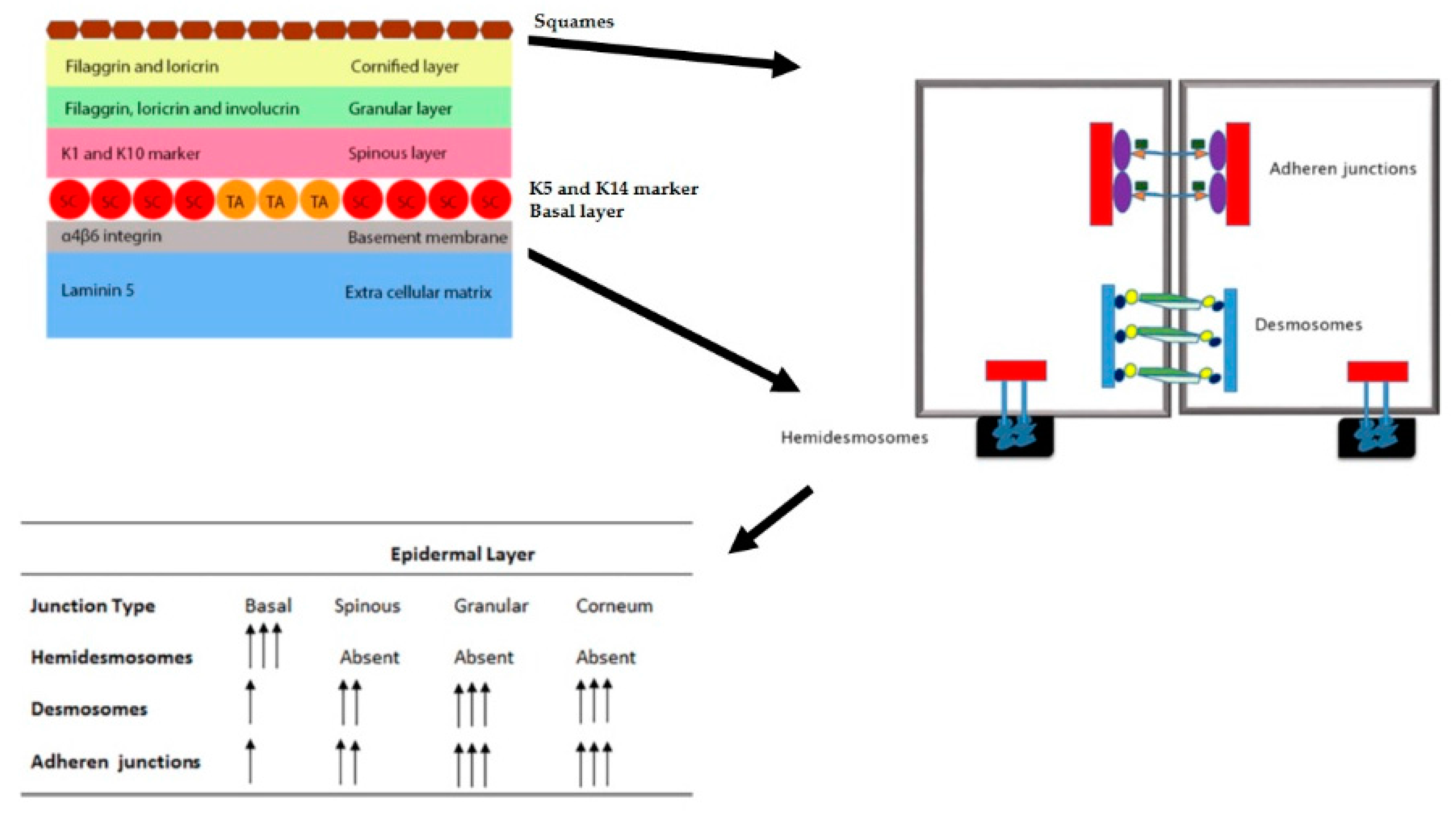
| Skin Layer | Intermediate Layers/Structures | Cell Populations Found | Type of Epidermal Stem Populations | Phenotypic Output of the Stem Cells | References |
|---|---|---|---|---|---|
| Epidermis | Basal layer | Undifferentiated cells | Interfollicular (IF) | IF epidermis | [1] |
| Spinous layer | Partially differentiated | – | – | [1] | |
| Granular layer | Partially differentiated | – | – | [1] | |
| Cornified layer | Terminally differentiated | – | – | [1] | |
| Bulge | Undifferentiated cells | Hair follicle stem cells (HFSC) | Hair follicle | [11] | |
| Sebaceous gland | Undifferentiated cells | sebaceous gland (SG) stem cells | Sebaceous gland | [12] |
2. Epidermal Stem Cells and Biomarkers
2.1. Integrins
2.2. Cadherins/Catenins
2.3. Keratins
2.4. p63
3. Epigenetic Regulation of Epidermal Stem Cell Biomarkers by miRNA Regulation
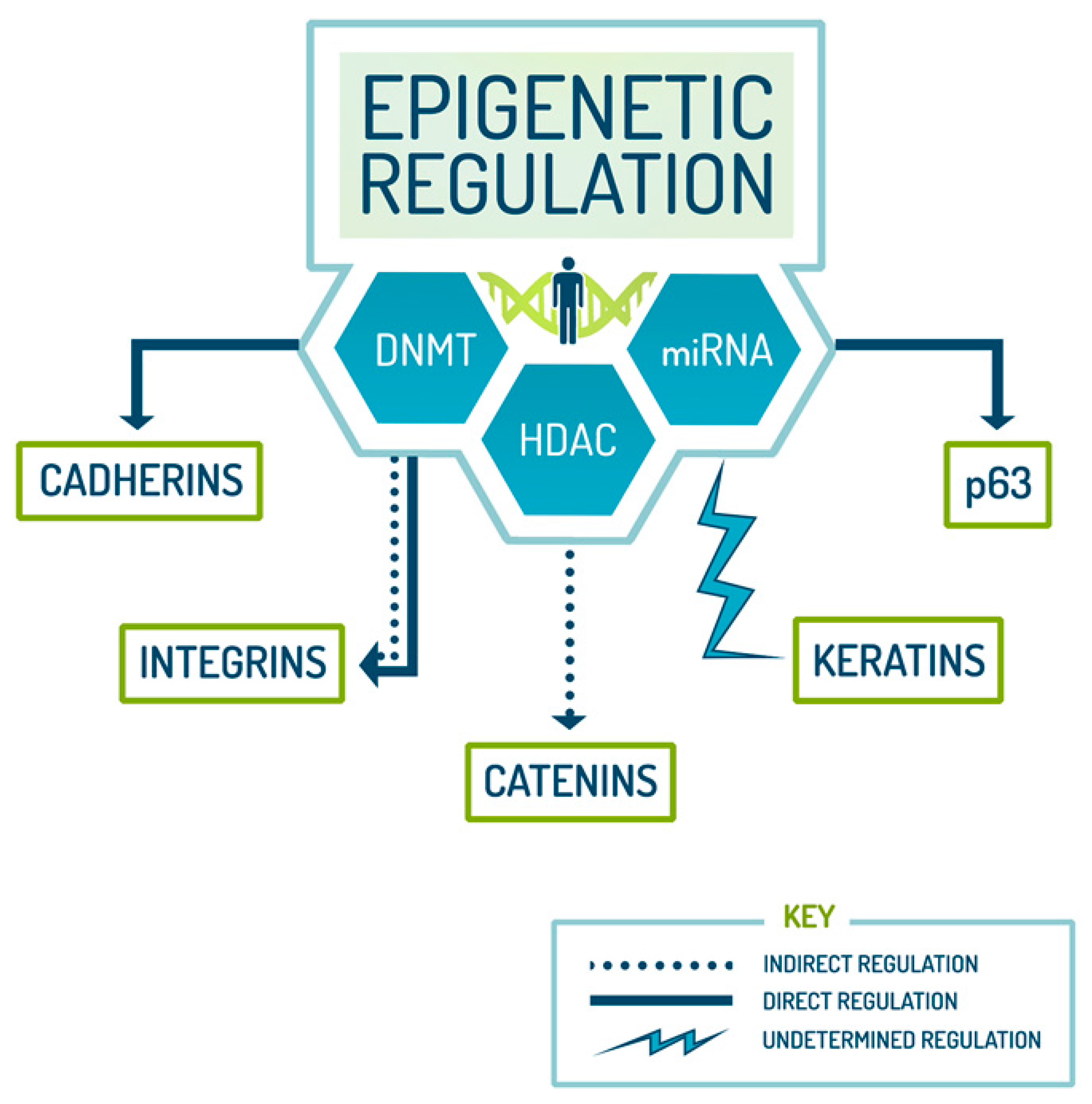
| Epigenetic Regulation of Stem Cells | ||
|---|---|---|
| Modifiers | Roles | References |
| miRNA | Stem cell differentiation and suppression of self-renewal, regulation of integrin, direct repression of p63 | [50,51,52] |
| DNMT | Stem cell differentiation and lineage commitment, cadherin regulation, catenin management | [53,54,55] |
| HDAC | Cell division/chromosome segregation accuracy, and pluripotency, cadherin regulation, p63 repression | [19,54,56] |
4. Epidermal Stem Cell (ESC) Biomarkers and Their Role in Wound Healing
4.1. Cadherins and Wound Healing
4.2. Integrins and Wound Healing
4.3. Catenins and Wound Healing
4.4. Keratins and Wound Healing
4.5. p63 and Wound Healing
5. Epigenetic Regulation of Wound Healing in Normal State and Disease
| Epigenetic modification | Enzyme involved | Epigenetic Effect | Effect on Epidermal development | Reference |
|---|---|---|---|---|
| Methylation | DNMT1 | Global hypermethylation | Maintenance of epidermal progenitor self-renewal capability | [108] |
| Histone methylation | Histone demethylase, JmjC domain-containing protein 3 (Jmjd3) | Demethylation of trimethylated histone H3 lysine 27 (H3K27me3) | Epidermal stratification, proliferation and differentiation | [19] |
| Ubiquitously transcribed X chromosome (UTX) | ||||
| Histone methyltransferase SET domain containing 8 (SETD8) | Histone H4 lysine 20 (H4K20) mono-methylation | – | [108] | |
| Histone acetylation | Histone deacetylase 1/2 | Global histone acetylation; H3 acetylation; P38 activation | Promote proliferation and differentiation of epidermal stem cells | [109] |
| Polycomb repressive complex 1 (PCR1) and polycomb repressive complex 2 (PCR2) | Enhancer of zeste 1 (Ezh1) and Enhancer of zeste 2 (Ezh2) | Trimethylation of histone H3 lysine 27 (H3K27) | Maintains stem cell quiescence regulates epidermal differentiation and stratification | [99,110] |
6. Conclusions
Acknowledgments
Authors Contributions
Conflicts of Interest
Abbreviations
References
- Blanpain, C.; Fuchs, E. Epidermal stem cells of the skin. Annu. Rev. Cell Dev. Biol. 2006, 22, 339–373. [Google Scholar] [CrossRef] [PubMed]
- Cichorek, M.; Wachulska, M.; Stasiewicz, A.; Tyminska, A. Skin melanocytes: Biology and development. Postep. Dermatol. Alergol. 2013, 30, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Senoo, M. Epidermal stem cells in homeostasis and wound repair of the skin. Adv. Wound Care (New Rochelle) 2013, 2, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Sotiropoulou, P.A.; Blanpain, C. Development and homeostasis of the skin epidermis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008383. [Google Scholar] [CrossRef] [PubMed]
- Eckert, R.L.; Rorke, E.A. Molecular biology of keratinocyte differentiation. Environ. Health Perspect. 1989, 80, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Blanpain, C.; Fuchs, E. Epidermal homeostasis: A balancing act of stem cells in the skin. Nat. Rev. Mol. Cell Biol. 2009, 10, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Waghmare, S.K.; Bansal, R.; Lee, J.; Zhang, Y.V.; McDermitt, D.J.; Tumbar, T. Quantitative proliferation dynamics and random chromosome segregation of hair follicle stem cells. EMBO J. 2008, 27, 1309–1320. [Google Scholar] [CrossRef] [PubMed]
- Morasso, M.I.; Tomic-Canic, M. Epidermal stem cells: The cradle of epidermal determination, differentiation and wound healing. Biol. Cell 2005, 97, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Ambler, C.A.; Maatta, A. Epidermal stem cells: Location, potential and contribution to cancer. J. Pathol. 2009, 217, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.C.; Li, L.; Fuchs, E. Transit-amplifying cells orchestrate stem cell activity and tissue regeneration. Cell 2014, 157, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lyle, S.; Yang, Z.; Cotsarelis, G. Keratin 15 promoter targets putative epithelial stem cells in the hair follicle bulge. J. Investig. Dermatol. 2003, 121, 963–968. [Google Scholar] [CrossRef] [PubMed]
- Niemann, C. Differentiation of the sebaceous gland. Derm. Endocrinol. 2009, 1, 64–67. [Google Scholar] [CrossRef]
- Breitkreutz, D.; Koxholt, I.; Thiemann, K.; Nischt, R. Skin basement membrane: The foundation of epidermal integrity—BM functions and diverse roles of bridging molecules nidogen and perlecan. Biomed. Res. Int. 2013, 2013, 179784. [Google Scholar] [CrossRef] [PubMed]
- Simpson, C.L.; Patel, D.M.; Green, K.J. Deconstructing the skin: Cytoarchitectural determinants of epidermal morphogenesis. Nat. Rev. Mol. Cell Biol. 2011, 12, 565–580. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Lee, J.B.; Kook, J.P.; Seo, J.J.; Nam, K.I.; Park, S.S.; Kim, Y.P. Expression of differentiation markers during fetal skin development in humans: Immunohistochemical studies on the precursor proteins forming the cornified cell envelope. J. Investig. Dermatol. 1999, 112, 882–886. [Google Scholar] [CrossRef] [PubMed]
- Sandilands, A.; Sutherland, C.; Irvine, A.D.; McLean, W.H. Filaggrin in the frontline: Role in skin barrier function and disease. J. Cell Sci. 2009, 122, 1285–1294. [Google Scholar] [CrossRef] [PubMed]
- Plikus, M.V.; Guerrero-Juarez, C.F.; Treffeisen, E.; Gay, D.L. Epigenetic control of skin and hair regeneration after wounding. Exp. Dermatol. 2015, 24, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, J.; Sen, C.K. MicroRNAs in skin and wound healing. Methods Mol. Biol. 2013, 936, 343–356. [Google Scholar] [PubMed]
- Lewis, C.J.; Mardaryev, A.N.; Sharov, A.A.; Fessing, M.Y.; Botchkarev, V.A. The epigenetic regulation of wound healing. Adv. Wound Care (New Rochelle) 2014, 3, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Eckert, R.L.; Adhikary, G.; Balasubramanian, S.; Rorke, E.A.; Vemuri, M.C.; Boucher, S.E.; Bickenbach, J.R.; Kerr, C. Biochemistry of epidermal stem cells. Biochim. Biophys. Acta 2013, 1830, 2427–2434. [Google Scholar] [CrossRef] [PubMed]
- Wray, H.; Mackenzie, I.C.; Storey, A.; Navsaria, H. α6 integrin and cd44 enrich for a primary keratinocyte population that displays resistance to UV-induced apoptosis. PLoS ONE 2012, 7, e46968. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, S.; Bauer, C.; Mundschau, G.; Li, Q.; Fuchs, E. Conditional ablation of β1 integrin in skin. Severe defects in epidermal proliferation, basement membrane formation, and hair follicle invagination. J. Cell Biol. 2000, 150, 1149–1160. [Google Scholar] [CrossRef] [PubMed]
- Tjin, M.S.; Chua, A.W.; Ma, D.R.; Lee, S.T.; Fong, E. Human epidermal keratinocyte cell response on integrin-specific artificial extracellular matrix proteins. Macromol. Biosci. 2014, 14, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Alitalo, K.; Kuismanen, E.; Myllyla, R.; Kiistala, U.; Asko-Seljavaara, S.; Vaheri, A. Extracellular matrix proteins of human epidermal keratinocytes and feeder 3T3 cells. J. Cell Biol. 1982, 94, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; de Paiva, C.S.; Luo, L.; Kretzer, F.L.; Pflugfelder, S.C.; Li, D.Q. Characterization of putative stem cell phenotype in human limbal epithelia. Stem Cells 2004, 22, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Raymond, K.; Kreft, M.; Janssen, H.; Calafat, J.; Sonnenberg, A. Keratinocytes display normal proliferation, survival and differentiation in conditional β4-integrin knockout mice. J. Cell Sci. 2005, 118, 1045–1060. [Google Scholar] [CrossRef] [PubMed]
- Paredes, J.; Figueiredo, J.; Albergaria, A.; Oliveira, P.; Carvalho, J.; Ribeiro, A.S.; Caldeira, J.; Costa, A.M.; Simoes-Correia, J.; Oliveira, M.J.; et al. Epithelial E- and P-cadherins: Role and clinical significance in cancer. Biochim. Biophys. Acta 2012, 1826, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Gumbiner, B.M. Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 2005, 6, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Tinkle, C.L.; Pasolli, H.A.; Stokes, N.; Fuchs, E. New insights into cadherin function in epidermal sheet formation and maintenance of tissue integrity. Proc. Natl. Acad. Sci. USA 2008, 105, 15405–15410. [Google Scholar] [CrossRef] [PubMed]
- Kobielak, A.; Boddupally, K. Junctions and inflammation in the skin. Cell Commun. Adhes. 2014, 21, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Lien, W.H.; Stepniak, E.; Vasioukhin, V. Dissecting the role of cadherin-catenin proteins in mammalian epidermis. Proc. Natl. Acad. Sci. USA 2008, 105, 15225–15226. [Google Scholar] [CrossRef] [PubMed]
- Ishiyama, N.; Ikura, M. The three-dimensional structure of the cadherin-catenin complex. Subcell. Biochem. 2012, 60, 39–62. [Google Scholar] [PubMed]
- Ishiyama, N.; Lee, S.H.; Liu, S.; Li, G.Y.; Smith, M.J.; Reichardt, L.F.; Ikura, M. Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion. Cell 2010, 141, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Stepniak, E.; Radice, G.L.; Vasioukhin, V. Adhesive and signaling functions of cadherins and catenins in vertebrate development. Cold Spring Harb. Perspect. Biol. 2009, 1, a002949. [Google Scholar] [CrossRef] [PubMed]
- Staunstrup, N.H.; Madsen, J.; Primo, M.N.; Li, J.; Liu, Y.; Kragh, P.M.; Li, R.; Schmidt, M.; Purup, S.; Dagnaes-Hansen, F.; et al. Development of transgenic cloned pig models of skin inflammation by DNA transposon-directed ectopic expression of human β1 and α2 integrin. PLoS ONE 2012, 7, e36658. [Google Scholar] [CrossRef] [PubMed]
- Garrod, D.; Chidgey, M. Desmosome structure, composition and function. Biochim. Biophys. Acta 2008, 1778, 572–587. [Google Scholar] [CrossRef] [PubMed]
- Deugnier, M.A.; Faraldo, M.M.; Rousselle, P.; Thiery, J.P.; Glukhova, M.A. Cell-extracellular matrix interactions and EGF are important regulators of the basal mammary epithelial cell phenotype. J. Cell Sci. 1999, 112, 1035–1044. [Google Scholar] [PubMed]
- Moles, J.P.; Watt, F.M. The epidermal stem cell compartment: Variation in expression levels of E-cadherin and catenins within the basal layer of human epidermis. J. Histochem. Cytochem. 1997, 45, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Kouklis, P.D.; Hutton, E.; Fuchs, E. Making a connection: Direct binding between keratin intermediate filaments and desmosomal proteins. J. Cell Biol. 1994, 127, 1049–1060. [Google Scholar] [CrossRef] [PubMed]
- Blanpain, C.; Lowry, W.E.; Pasolli, H.A.; Fuchs, E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 2006, 20, 3022–3035. [Google Scholar] [CrossRef] [PubMed]
- Bose, A.; Teh, M.T.; Hutchison, I.L.; Wan, H.; Leigh, I.M.; Waseem, A. Two mechanisms regulate keratin k15 expression in keratinocytes: Role of PKC/AP-1 and FOXM1 mediated signalling. PLoS ONE 2012, 7, e38599. [Google Scholar] [CrossRef] [PubMed]
- Bose, A.; Teh, M.T.; Mackenzie, I.C.; Waseem, A. Keratin k15 as a biomarker of epidermal stem cells. Int. J. Mol. Sci. 2013, 14, 19385–19398. [Google Scholar] [CrossRef] [PubMed]
- Lane, E.B.; McLean, W.H. Keratins and skin disorders. J. Pathol. 2004, 204, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Calabro, V.; Mansueto, G.; Santoro, R.; Gentilella, A.; Pollice, A.; Ghioni, P.; Guerrini, L.; La Mantia, G. Inhibition of p63 transcriptional activity by p14ARF: Functional and physical link between human ARF tumor suppressor and a member of the p53 family. Mol. Cell. Biol. 2004, 24, 8529–8540. [Google Scholar] [CrossRef] [PubMed]
- Koster, M.I.; Kim, S.; Mills, A.A.; DeMayo, F.J.; Roop, D.R. P63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004, 18, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Ying, H.; Chang, D.L.; Zheng, H.; McKeon, F.; Xiao, Z.X. DNA-binding and transactivation activities are essential for TAp63 protein degradation. Mol. Cell. Biol. 2005, 25, 6154–6164. [Google Scholar] [CrossRef] [PubMed]
- Koster, M.I.; Dai, D.; Roop, D.R. Conflicting roles for p63 in skin development and carcinogenesis. Cell Cycle 2007, 6, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Truong, A.B.; Kretz, M.; Ridky, T.W.; Kimmel, R.; Khavari, P.A. P63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006, 20, 3185–3197. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M. Epigenetic inheritance and the missing heritability problem. Genetics 2009, 182, 845–850. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Fuchs, E. MicroRNAs and their roles in mammalian stem cells. J. Cell Sci. 2011, 124, 1775–1783. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Song, J.; Han, J.; Kim, Y.; Chun, C.H.; Jin, E.J. Two non-coding RNAs, microRNA-101 and hottip contribute cartilage integrity by epigenetic and homeotic regulation of integrin-α1. Cell Signal. 2013, 25, 2878–2887. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Poy, M.N.; Stoffel, M.; Fuchs, E. A skin microRNA promotes differentiation by repressing “stemness”. Nature 2008, 452, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Kim, J.; Yuan, X.; Braun, T. Epigenetic modifications of stem cells: A paradigm for the control of cardiac progenitor cells. Circ. Res. 2011, 109, 1067–1081. [Google Scholar] [CrossRef] [PubMed]
- Yi, T.Z.; Li, J.; Han, X.; Guo, J.; Qu, Q.; Guo, L.; Sun, H.D.; Tan, W.H. DNMT inhibitors and HDAC inhibitors regulate E-cadherin and Bcl-2 expression in endometrial carcinoma in vitro and in vivo. Chemotherapy 2012, 58, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Jamaladdin, S.; Kelly, R.D.; O’Regan, L.; Dovey, O.M.; Hodson, G.E.; Millard, C.J.; Portolano, N.; Fry, A.M.; Schwabe, J.W.; Cowley, S.M. Histone deacetylase (HDAC) 1 and 2 are essential for accurate cell division and the pluripotency of embryonic stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, 9840–9845. [Google Scholar] [CrossRef] [PubMed]
- Savickiene, J.; Treigyte, G.; Jazdauskaite, A.; Borutinskaite, V.V.; Navakauskiene, R. DNA methyltransferase inhibitor RG108 and histone deacetylase inhibitors cooperate to enhance NB4 cell differentiation and E-cadherin re-expression by chromatin remodelling. Cell Biol. Int. 2012, 36, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Nelson, W.J.; Dickinson, D.J.; Weis, W.I. Roles of cadherins and catenins in cell-cell adhesion and epithelial cell polarity. Prog. Mol. Biol. Transl. Sci. 2013, 116, 3–23. [Google Scholar] [PubMed]
- Takada, Y.; Ye, X.; Simon, S. The integrins. Genome Biol. 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- Naganuma, K.; Hatta, M.; Ikebe, T.; Yamazaki, J. Epigenetic alterations of the keratin 13 gene in oral squamous cell carcinoma. BMC Cancer 2014, 14. [Google Scholar] [CrossRef] [PubMed]
- Sethi, I.; Sinha, S.; Buck, M.J. Role of chromatin and transcriptional co-regulators in mediating p63-genome interactions in keratinocytes. BMC Genom. 2014, 15, 1042. [Google Scholar] [CrossRef] [PubMed]
- Botchkarev, V.A.; Gdula, M.R.; Mardaryev, A.N.; Sharov, A.A.; Fessing, M.Y. Epigenetic regulation of gene expression in keratinocytes. J. Investig. Dermatol. 2012, 132, 2505–2521. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Bartel, D.P.; Chen, C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004, 5, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, J.; Zhu, Y.; Dai, Y.; Zeng, T.; Liu, L.; Li, J.; Wang, H.; Qin, Y.; Zeng, M.; et al. MicroRNA-9 promotes tumor metastasis via repressing E-cadherin in esophageal squamous cell carcinoma. Oncotarget 2014, 5, 11669–11680. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Zhang, A.; Shi, Z.; Ma, F.; Pu, P.; Wang, T.; Zhang, J.; Kang, C.; Zhang, Q. MicroRNA-200a suppresses the Wnt/β-catenin signaling pathway by interacting with β-catenin. Int. J. Oncol. 2012, 40, 1162–1170. [Google Scholar] [PubMed]
- Yamada, N.; Noguchi, S.; Mori, T.; Naoe, T.; Maruo, K.; Akao, Y. Tumor-suppressive microRNA-145 targets catenin δ-1 to regulate Wnt/β-catenin signaling in human colon cancer cells. Cancer Lett. 2013, 335, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Hirata, H.; Hinoda, Y.; Ueno, K.; Nakajima, K.; Ishii, N.; Dahiya, R. MicroRNA-1826 directly targets β-catenin (CTNNB1) and MEK1 (MAP2K1) in VHL-inactivated renal cancer. Carcinogenesis 2012, 33, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Hirata, H.; Hinoda, Y.; Ueno, K.; Shahryari, V.; Tabatabai, Z.L.; Dahiya, R. MicroRNA-1826 targets VEGFC, β-catenin (CTNNB1) and MEK1 (MAP2K1) in human bladder cancer. Carcinogenesis 2012, 33, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Von Frowein, J.; Pagel, P.; Kappler, R.; von Schweinitz, D.; Roscher, A.; Schmid, I. MicroRNA-492 is processed from the keratin 19 gene and up-regulated in metastatic hepatoblastoma. Hepatology 2011, 53, 833–842. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Harbeck, M.C.; Zhang, W.; Jacobson, J.R. MicroRNA regulation of integrins. Transl. Res. 2013, 162, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.H.; Huang, W.C.; Chang, J.W.; Chang, K.J.; Kuo, W.H.; Wang, M.Y.; Lin, K.Y.; Uen, Y.H.; Hou, M.F.; Lin, C.M.; et al. MicroRNA-149 targets GIT1 to suppress integrin signaling and breast cancer metastasis. Oncogene 2014, 33, 4496–4507. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chang, L.; Du, W.W.; Gupta, S.; Khorshidi, A.; Sefton, M.; Yang, B.B. Anti-microRNA-378a enhances wound healing process by upregulating integrin β-3 and vimentin. Mol. Ther. 2014, 22, 1839–1850. [Google Scholar] [CrossRef] [PubMed]
- Sivasankar, S. Tuning the kinetics of cadherin adhesion. J. Investig. Dermatol. 2013, 133, 2318–2323. [Google Scholar] [CrossRef] [PubMed]
- Maxson, S.; Lopez, E.A.; Yoo, D.; Danilkovitch-Miagkova, A.; Leroux, M.A. Concise review: Role of mesenchymal stem cells in wound repair. Stem Cells Transl. Med. 2012, 1, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Colonna, M. Cytolytic responses: Cadherins put out the fire. J. Exp. Med. 2006, 203, 261–264. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Bhattacharya, S.; Bajaj, G.; Guha, G.; Wang, Z.; Jang, H.S.; Leid, M.; Indra, A.K.; Ganguli-Indra, G. Delayed cutaneous wound healing and aberrant expression of hair follicle stem cell markers in mice selectively lacking Ctip2 in epidermis. PLoS ONE 2012, 7, e29999. [Google Scholar] [CrossRef] [PubMed]
- Keswell, D.; Kidson, S.H.; Davids, L.M. Melanocyte migration is influenced by E-cadherin-dependent adhesion of keratinocytes in both two- and three-dimensional in vitro wound models. Cell Biol. Int. 2015, 39, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Longmate, W.M.; Dipersio, C.M. Integrin regulation of epidermal functions in wounds. Adv. Wound Care 2014, 3, 229–246. [Google Scholar] [CrossRef] [PubMed]
- Gagen, D.; Faralli, J.A.; Filla, M.S.; Peters, D.M. The role of integrins in the trabecular meshwork. J. Ocul. Pharmacol. Ther. 2014, 30, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Israeli-Rosenberg, S.; Manso, A.M.; Okada, H.; Ross, R.S. Integrins and integrin-associated proteins in the cardiac myocyte. Circ. Res. 2014, 114, 572–586. [Google Scholar] [CrossRef] [PubMed]
- Koivisto, L.; Heino, J.; Hakkinen, L.; Larjava, H. Integrins in wound healing. Adv. Wound Care 2014, 3, 762–783. [Google Scholar] [CrossRef] [PubMed]
- Drees, F.; Pokutta, S.; Yamada, S.; Nelson, W.J.; Weis, W.I. α-Catenin is a molecular switch that binds E-cadherin-β-catenin and regulates actin-filament assembly. Cell 2005, 123, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Bielefeld, K.A.; Amini-Nik, S.; Alman, B.A. Cutaneous wound healing: Recruiting developmental pathways for regeneration. Cell. Mol. Life Sci. 2013, 70, 2059–2081. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Watanabe, K.; Fallahi, M.; Lee, B.; Afetian, M.E.; Rheaume, C.; Wu, D.; Horsley, V.; Dai, X. Pygo2 regulates β-catenin-induced activation of hair follicle stem/progenitor cells and skin hyperplasia. Proc. Natl. Acad. Sci. USA 2014, 111, 10215–10220. [Google Scholar] [CrossRef] [PubMed]
- Stojadinovic, O.; Brem, H.; Vouthounis, C.; Lee, B.; Fallon, J.; Stallcup, M.; Merchant, A.; Galiano, R.D.; Tomic-Canic, M. Molecular pathogenesis of chronic wounds: The role of β-catenin and c-myc in the inhibition of epithelialization and wound healing. Am. J. Pathol. 2005, 167, 59–69. [Google Scholar] [CrossRef]
- Patel, G.K.; Wilson, C.H.; Harding, K.G.; Finlay, A.Y.; Bowden, P.E. Numerous keratinocyte subtypes involved in wound re-epithelialization. J. Investig. Dermatol. 2006, 126, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Pechter, P.M.; Gil, J.; Valdes, J.; Tomic-Canic, M.; Pastar, I.; Stojadinovic, O.; Kirsner, R.S.; Davis, S.C. Keratin dressings speed epithelialization of deep partial-thickness wounds. Wound Repair Regen. 2012, 20, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Than, M.P.; Smith, R.A.; Hammond, C.; Kelly, R.; Marsh, C.; Maderal, A.D.; Kirsner, R.S. Keratin-based wound care products for treatment of resistant vascular wounds. J. Clin. Aesthet. Dermatol. 2012, 5, 31–35. [Google Scholar] [PubMed]
- Loschke, F.; Homberg, M.; Magin, T.M. Keratin isotypes control desmosome stability and dynamics through PKCα. J. Investig. Dermatol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, D.; Senoo, M. Expansion of epidermal progenitors with high p63 phosphorylation during wound healing of mouse epidermis. Exp. Dermatol. 2013, 22, 374–376. [Google Scholar] [CrossRef] [PubMed]
- Botchkarev, V.A.; Flores, E.R. P53/p63/p73 in the epidermis in health and disease. Cold Spring Harb. Perspect. Med. 2014, 4, a015248. [Google Scholar] [CrossRef] [PubMed]
- Warner, S.M.; Hackett, T.L.; Shaheen, F.; Hallstrand, T.S.; Kicic, A.; Stick, S.M.; Knight, D.A. Transcription factor p63 regulates key genes and wound repair in human airway epithelial basal cells. Am. J. Respir. Cell Mol. Biol. 2013, 49, 978–988. [Google Scholar] [CrossRef] [PubMed]
- Eming, S.A.; Krieg, T.; Davidson, J.M. Inflammation in wound repair: Molecular and cellular mechanisms. J. Investig. Dermatol. 2007, 127, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.C.; Li, L.; Fuchs, E. Emerging interactions between skin stem cells and their niches. Nat. Med. 2014, 20, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Duan, E. Epigenetic regulations on skin wound healing: Implications from current researches. Ann. Transl. Med. 2015, 3, 227. [Google Scholar] [PubMed]
- Ti, D.; Li, M.; Fu, X.; Han, W. Causes and consequences of epigenetic regulation in wound healing. Wound Repair Regen. 2014, 22, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Mann, J.; Mann, D.A. Epigenetic regulation of wound healing and fibrosis. Curr. Opin. Rheumatol. 2013, 25, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Jin, H.; Wang, X. Epidermal stem cells and their epigenetic regulation. Int. J. Mol. Sci. 2013, 14, 17861–17880. [Google Scholar] [CrossRef] [PubMed]
- Stroncek, J.D.; Reichert, W.M. Overview of wound healing in different tissue types. In Indwelling Neural Implants: Strategies for Contending with the in Vivo Environment; Reichert, W.M., Ed.; CRC Press/Taylor & Francis: Boca Raton, FL, USA, 2008. [Google Scholar]
- Sanchis, A.; Alba, L.; Latorre, V.; Sevilla, L.M.; Perez, P. Keratinocyte-targeted overexpression of the glucocorticoid receptor delays cutaneous wound healing. PLoS ONE 2012, 7, e29701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thandavarayan, R.A.; Garikipati, V.N.; Joladarashi, D.; Suresh Babu, S.; Jeyabal, P.; Verma, S.K.; Mackie, A.R.; Khan, M.; Arumugam, S.; Watanabe, K.; et al. Sirtuin-6 deficiency exacerbates diabetes-induced impairment of wound healing. Exp. Dermatol. 2015, 24, 773–778. [Google Scholar] [CrossRef] [PubMed]
- Spallotta, F.; Cencioni, C.; Straino, S.; Sbardella, G.; Castellano, S.; Capogrossi, M.C.; Martelli, F.; Gaetano, C. Enhancement of lysine acetylation accelerates wound repair. Commun. Integr. Biol. 2013, 6, e25466. [Google Scholar] [CrossRef] [PubMed]
- Komine, M.; Rao, L.S.; Freedberg, I.M.; Simon, M.; Milisavljevic, V.; Blumenberg, M. Interleukin-1 induces transcription of keratin k6 in human epidermal keratinocytes. J. Investig. Dermatol. 2001, 116, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Coulombe, P.A. Defining a region of the human keratin 6a gene that confers inducible expression in stratified epithelia of transgenic mice. J. Biol. Chem. 1997, 272, 11979–11985. [Google Scholar] [CrossRef] [PubMed]
- Wawersik, M.; Paladini, R.D.; Noensie, E.; Coulombe, P.A. A proline residue in the α-helical rod domain of type I keratin 16 destabilizes keratin heterotetramers. J. Biol. Chem. 1997, 272, 32557–32565. [Google Scholar] [CrossRef] [PubMed]
- Wong, P.; Coulombe, P.A. Loss of keratin 6 (k6) proteins reveals a function for intermediate filaments during wound repair. J. Cell Biol. 2003, 163, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Sen, G.L.; Reuter, J.A.; Webster, D.E.; Zhu, L.; Khavari, P.A. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 2010, 463, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Connelly, J.T.; Mishra, A.; Gautrot, J.E.; Watt, F.M. Shape-induced terminal differentiation of human epidermal stem cells requires p38 and is regulated by histone acetylation. PLoS ONE 2011, 6, e27259. [Google Scholar] [CrossRef] [PubMed]
- Ezhkova, E.; Lien, W.H.; Stokes, N.; Pasolli, H.A.; Silva, J.M.; Fuchs, E. Ezh1 and Ezh2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011, 25, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Petersson, M.; Brylka, H.; Kraus, A.; John, S.; Rappl, G.; Schettina, P.; Niemann, C. TCF/Lef1 activity controls establishment of diverse stem and progenitor cell compartments in mouse epidermis. EMBO J. 2011, 30, 3004–3018. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, D.; Senoo, M. Increased p63 phosphorylation marks early transition of epidermal stem cells to progenitors. J. Investig. Dermatol. 2012, 132, 2461–2464. [Google Scholar] [CrossRef] [PubMed]
- Bale, A.E.; Yu, K.P. The hedgehog pathway and basal cell carcinomas. Hum. Mol. Genet. 2001, 10, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Abidi, A. Hedgehog signaling pathway: A novel target for cancer therapy: Vismodegib, a promising therapeutic option in treatment of basal cell carcinomas. Indian J. Pharmacol. 2014, 46, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Y.; Wang, J.; Mancianti, M.L.; Epstein, E.H., Jr. Basal cell carcinomas arise from hair follicle stem cells in Ptch1+/− mice. Cancer Cell 2011, 19, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.Y.; Reiter, J.F. Wounding mobilizes hair follicle stem cells to form tumors. Proc. Natl. Acad. Sci. USA 2011, 108, 4093–4098. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saldanha, S.N.; Royston, K.J.; Udayakumar, N.; Tollefsbol, T.O. Epigenetic Regulation of Epidermal Stem Cell Biomarkers and Their Role in Wound Healing. Int. J. Mol. Sci. 2016, 17, 16. https://doi.org/10.3390/ijms17010016
Saldanha SN, Royston KJ, Udayakumar N, Tollefsbol TO. Epigenetic Regulation of Epidermal Stem Cell Biomarkers and Their Role in Wound Healing. International Journal of Molecular Sciences. 2016; 17(1):16. https://doi.org/10.3390/ijms17010016
Chicago/Turabian StyleSaldanha, Sabita N., Kendra J. Royston, Neha Udayakumar, and Trygve O. Tollefsbol. 2016. "Epigenetic Regulation of Epidermal Stem Cell Biomarkers and Their Role in Wound Healing" International Journal of Molecular Sciences 17, no. 1: 16. https://doi.org/10.3390/ijms17010016
APA StyleSaldanha, S. N., Royston, K. J., Udayakumar, N., & Tollefsbol, T. O. (2016). Epigenetic Regulation of Epidermal Stem Cell Biomarkers and Their Role in Wound Healing. International Journal of Molecular Sciences, 17(1), 16. https://doi.org/10.3390/ijms17010016