Ile-1781-Leu and Asp-2078-Gly Mutations in ACCase Gene, Endow Cross-resistance to APP, CHD, and PPZ in Phalaris minor from Mexico
Abstract
:1. Introduction
2. Results
2.1. Dose-Response
| Herbicide | Biotype | d | c | b | R2 | P | GR50 (g ai ha−1) | RF |
|---|---|---|---|---|---|---|---|---|
| Fenoxaprop-p-ethyl | S | 99.73 | 11.42 | 4.39 | 0.99 | <0.0001 | 25.62 | 12.32 |
| R | 100.00 | 8.94 | 4.29 | 0.94 | <0.0001 | 315.40 | ||
| Cyhalofop-butyl | S | 100.00 | 8.78 | 1.02 | 0.96 | <0.0001 | 31.94 | 22.58 |
| R | 100.00 | 30.71 | 2.12 | 0.97 | <0.0001 | 721.32 | ||
| Diclofop-p-methyl | S | 100.00 | 15.26 | 1.94 | 0.94 | <0.0001 | 140.00 | 9.00 |
| R | 100.00 | 26.13 | 2.77 | 0.93 | <0.0001 | 1260.00 | ||
| Propaquizafop | S | 100.00 | 10.54 | 3.21 | 0.99 | <0.0001 | 28.44 | 12.58 |
| R | 100.00 | 32.07 | 3.06 | 0.92 | <0.0001 | 357.43 | ||
| Clethodim | S | 100.00 | 0.00 | 1.85 | 0.91 | <0.0001 | 9.37 | 5.03 |
| R | 100.00 | 0.00 | 1.46 | 0.94 | <0.0001 | 46.81 | ||
| Cycloxydim | S | 100.00 | 0.00 | 2.38 | 0.94 | <0.0001 | 11.01 | 19.07 |
| R | 100.00 | 0.00 | 1.03 | 0.92 | <0.0001 | 210.00 | ||
| Tralkoxydim | S | 100.00 | 0.00 | 5.49 | 0.99 | <0.0001 | 214.75 | 2.05 |
| R | 100.00 | 17.21 | 3.53 | 0.98 | <0.0001 | 442.00 | ||
| Pinoxaden | S | 99.86 | 0.00 | 1.18 | 0.99 | <0.0001 | 7.32 | 14.80 |
| R | 100.00 | 0.00 | 2.03 | 0.99 | <0.0001 | 108.39 |
2.2. [14C]Diclofop-Methyl(DM) Uptake and Translocation
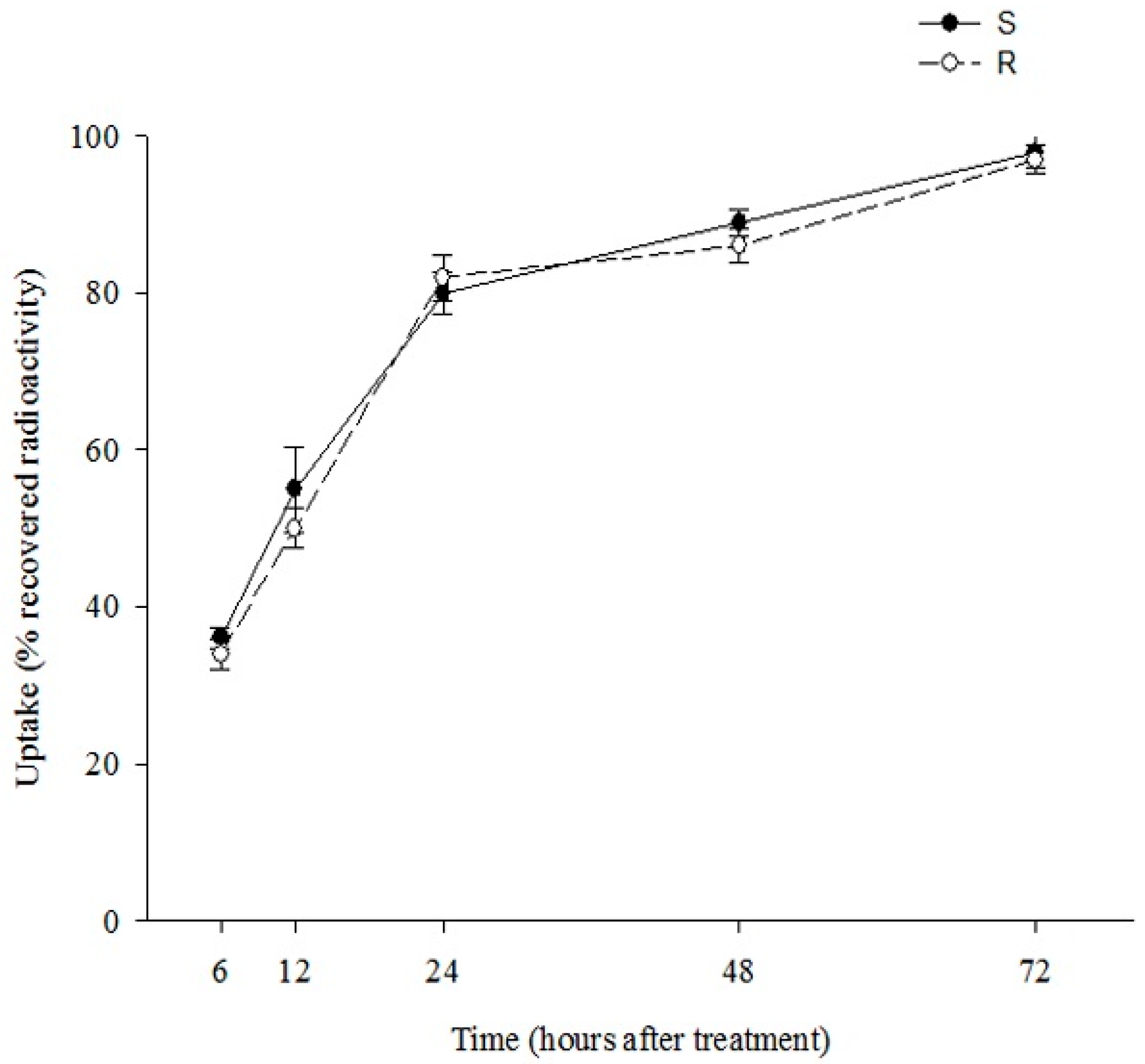
| % Absorption | Translocation (% of Absorbed) a | |||
|---|---|---|---|---|
| Treated Leaf | Rest of Plant | Roots | ||
| S | 94.0 ± 2.01 a | 94.6 ± 2.78 a | 5.4 ± 1.23 b | nd |
| R | 99.3 ± 3.54 a | 93.9 ± 3.66 a | 6.1 ± 3.26 b | nd |
2.3. Metabolic Studies Using [14C]Diclofop-Methyl (DM)
| Components | % Extracted Radioactivity a | |
|---|---|---|
| R | S | |
| DM | 48.1 ± 3.94 a | 46.3 ± 3.21 a |
| Diclofop | 34.6 ± 2.78 b | 35.8 ± 4.79 b |
| Conjugates | 17.8 ± 4.33 c | 16.4 ± 5.63 c |
2.4. Enzyme Activity
| Herbicide | Biotype | d | c | b | R2 | p | I50 (µM) | RF |
|---|---|---|---|---|---|---|---|---|
| Fenoxaprop-p-ethyl | S | 100.05 | 0.00 | 1.25 | 0.94 | <0.0001 | 0.53 | 14.81 |
| R | 100.00 | 0.05 | 1.87 | 0.96 | <0.0001 | 7.85 | ||
| Cyhalofop-butyl | S | 100.87 | 0.16 | 0.94 | 0.93 | <0.0001 | 8.90 | 8.72 |
| R | 100.61 | 12.48 | 0.65 | 0.97 | <0.0001 | 77.68 | ||
| Diclofop-p-methyl | S | 100.59 | 0.08 | 0.78 | 0.93 | <0.0001 | 0.62 | 11.51 |
| R | 100.41 | 0.78 | 1.51 | 0.98 | <0.0001 | 7.14 | ||
| Setoxydim | S | 100.68 | 23.00 | 1.23 | 0.96 | <0.0001 | 584.87 | 2.56 |
| R | 100.77 | 44.34 | 0.29 | 0.97 | <0.0001 | 1500.76 | ||
| Tralkoxydim | S | 100.39 | 1.35 | 1.64 | 0.92 | <0.0001 | 1.68 | 5.24 |
| R | 101.87 | 0.54 | 1.18 | 0.91 | <0.0001 | 8.81 | ||
| Pinoxaden | S | 99.87 | 0.42 | 0.78 | 0.98 | <0.0001 | 0.39 | 23.41 |
| R | 99.72 | 9.27 | 1.23 | 0.96 | <0.0001 | 9.13 |
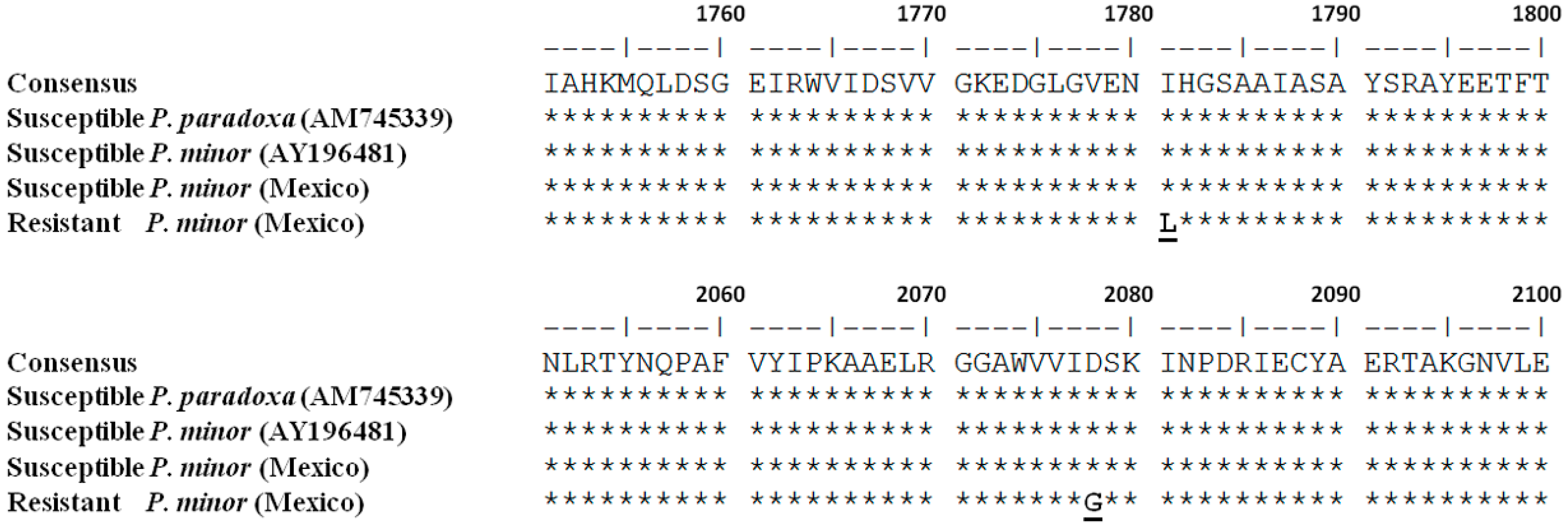
2.5. Molecular Analysis
3. Discussion
4. Experimental Section
4.1. Chemicals
4.2. Plant Materials
4.3. Growth Assays
4.4. Dose-Response Assays
| Herbicide | Rate (g ai ha−1) | |
|---|---|---|
| Biotype S | Biotype R | |
| Clethodim | 0, 40, 80, 100, 150, 200 | 0, 80, 100, 150, 200, 400 |
| Cyhalofop-butyl | 0, 20, 30, 40, 50, 70 | 0, 100, 300, 600, 700, 900 |
| Tralkoxydim | 0, 200, 300, 400, 500 | 0, 300, 400, 500, 600 |
| Fenoxaprop-p-ethyl | 0, 20, 30, 40, 45, 50 | 0, 200, 300, 400, 500, 600 |
| Propaquizafop | 0, 20, 40, 60, 100 | 0, 3000, 3500, 4000, 5000 |
| Diclofop-methyl | 0, 72, 144, 216, 360 | 0, 3000, 3500, 4000, 5000 |
| Cycloxidim | 0, 20, 40, 60, 100 | 0, 2000, 3000, 3500, 4000 |
| Pinoxaden | 0, 4, 8, 16, 32, 64 | 0, 25, 50, 100, 200, 400 |
4.5. Enzyme Purification and ACCase Activity Assay
4.6. Foliar Uptake and Translocation of [14C]Diclofop-Methyl (DM)
4.7. Phosphor Imaging
4.8. Metabolic Study with [14C]Diclofop-Methyl (DM)
4.9. Molecular Analysis
4.9.1. DNA Extraction and PCR Amplification
5. Conclusions
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Cruz-Hipólito, H.E. Gramíneas Resistentes a Herbicidas en Latinoamérica: Aspectos Agronómicos, Bioquímicos y Moleculares. Ph.D. Thesis, University of Cordoba, Córdoba, Spain, 2010. [Google Scholar]
- Weber, E.; Gut, D. Assessing the risk of potentially invasive plant species in central Europe. J. Nat. Conserv. 2004, 12, 171–179. [Google Scholar] [CrossRef]
- Shaner, D.L. Herbicide Handbook, 10th ed.; Weed Science Society of America: Lawrence, KS, USA, 2014. [Google Scholar]
- Hochberg, O.; Sibony, M.; Rubin, B. The response of ACCase-resistant Phalaris paradoxa populations involves two different target site mutations. Weed Res. 2009, 49, 37–46. [Google Scholar] [CrossRef]
- Devine, D.M.; Shimabukuro, R.H. Resistance to acetyl coenzyme A carboxylase inhibiting herbicides. In Herbicide Resistance in Plants; Powles, S.B., Holtum, J.A.M., Eds.; CRC Press: Boca Raton, FL, USA, 1994; pp. 141–169. [Google Scholar]
- Gherekhloo, J.; Rashed-Mohassel, M.H.; Mahalati, M.N.; Zand, E.; Ghanbari, A.; Osuna, M.D.; De Prado, R. Confirmed resistance to aryloxyphenoxypropionate herbicides in Phalaris minor populations in Iran. Weed Biol. Manag. 2011, 11, 29–37. [Google Scholar] [CrossRef]
- Collavo, A.; Panozzo, S.; Lucchesi, G.; Scarabel, L.; Sattin, M. Characterization and management of Phalaris paradoxa resistant to ACCase-inhibitors. Crop Prot. 2011, 30, 293–299. [Google Scholar] [CrossRef]
- Heap, I. The International Survey of Herbicide Resistant Weeds. Available online: http://www.weedscience.com (accessed on 10 July 2015).
- De Prado, J.L.; Osuna, M.D.; Heredia, A.; De Prado, R. Lolium rigidum, a pool of resistance mechanisms to ACCase inhibitor herbicides. J. Agric. Food Chem. 2005, 53, 2185–2191. [Google Scholar] [CrossRef] [PubMed]
- Délye, C.; Pernin, F.; Michel, S. ‘Universal’ PCR assays detecting mutations in acetyl-coenzyme A carboxylase or acetolactate synthase that endow herbicide resistance in grass weeds. Weed Res. 2011, 51, 353–362. [Google Scholar] [CrossRef]
- Shukla, A.; Dupont, S.; Devine, M.D. Resistance to ACCase inhibitor herbicides in wild oat: Evidence for target site-based resistance in two biotypes from Canada. Pestic. Biochem. Physiol. 1997, 57, 147–155. [Google Scholar] [CrossRef]
- Seefeldt, S.S.; Fuerst, E.P.; Gealy, D.R.; Shukla, A.; Irzyk, G.P.; Devine, M.D. Mechanisms of resistance to diclofop of two wild-oat (Avena fatua) biotypes from the Williamette Valley of Oregon. Weed Sci. 1996, 44, 776–781. [Google Scholar]
- Shimabukuro, R.H.; Hoffer, B.L. Effect of diclofop on the membrane potentials of herbicide-resistant and susceptible annual ryegrass root tips. Plant Physiol. 1992, 98, 1415–1422. [Google Scholar] [CrossRef] [PubMed]
- De Prado, J.L.; De Prado, R.; Shimabukuro, R.H. The effect of diclofop on membrane potential, ethylene induction, and herbicide phytotoxicity in resistant and susceptible biotypes of grasses. Pestic. Biochem. Phys. 1999, 63, 1–14. [Google Scholar] [CrossRef]
- Ahmad-Hamdani, M.S.; Yu, Q.; Han, H.; Cawthray, G.R.; Wang, S.F.; Powles, S.B. Herbicide resistance endowed by enhanced rates of herbicide metabolism in wild oat (Avena spp.). Weed Sci. 2013, 61, 55–62. [Google Scholar] [CrossRef]
- Powles, S.B.; Yu, Q. Evolution in action: Plants resistant to herbicides. Annu. Rev. Plant Biol. 2010, 61, 317–347. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Hipolito, H.; Osuna, M.D.; Domínguez-Valenzuela, J.A.; Espinoza, N.; de Prado, R. Mechanism of resistance to ACCase-inhibiting herbicides in wild oat (Avena fatua) from Latin America. J. Agric. Food Chem. 2011, 59, 7261–7267. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Harrison, D.K.; Chalupska, D.; Gornicki, P.; O’Donnell, C.C.; Adkins, S.W.; Haselkorn, R.; Williams, R.R. Single-site mutations in the carboxyl transferase domain of plastid acetyl-CoA carboxylase confer resistance to grass-specific herbicides. Proc. Natl. Acad. Sci. USA 2007, 104, 3627–3632. [Google Scholar] [CrossRef] [PubMed]
- Kaundun, S.S.; Hutchings, S.J.; Dale, R.P.; McIndoe, E. Role of a novel I1781T mutation and other mechanisms in conferring resistance to Acetyl-CoA carboxylase inhibiting herbicides in a black-grass population. PLoS ONE 2013, 8, e69568. [Google Scholar] [CrossRef] [PubMed]
- Gherekhloo, J.; Osuna, M.D.; De Prado, R. Biochemical and molecular basis of resistance to ACCase-inhibiting herbicides in Iranian Phalaris minor populations. Weed Res. 2012, 52, 367–372. [Google Scholar] [CrossRef]
- Kaundun, S.S.; Hutchings, S.J.; Dale, R.P.; McIndoe, E. Broad resistance to ACCase inhibiting herbicides in a ryegrass population is due only to a cysteine to arginine mutation in the target enzyme. PLoS ONE 2012, 7, e39759. [Google Scholar] [CrossRef] [PubMed]
- Heap, I.; Murray, B.G.; Loeppky, H.A.; Morrison, I.N. Resistance to aryloxyphenoxy propionate and cyclohexanedione herbicides in wild oat (Avena fatua). Weed Sci. 1993, 41, 232–238. [Google Scholar]
- Osuna, M.D.; Goulart, I.C.G.R.; Vidal, R.A.; Kalsing, A.; Ruiz-Santaella, L.P.; de Prado, R. Resistance to ACCase inhibitors in Eleusine indica from Brazil involves a target site mutation. Planta Daninha 2012, 30, 675–681. [Google Scholar] [CrossRef]
- Délye, C.; Matéjicek, A.; Michel, S. Cross-resistance patterns to ACCase-inhibiting herbicides conferred by mutant ACCase isoforms in Alopecurus myosuroides Huds. (black-grass), re-examined at the recommended herbicide field rate. Pest Manag. Sci. 2008, 64, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Hipolito, H.; Domínguez-Valenzuela, J.; Osuna, M.D.; De Prado, R. Resistance mechanism to acetyl coenzyme A carboxylase inhibiting herbicides in Phalaris paradoxa collected in Mexican wheat fields. Plant Soil 2012, 355, 121–130. [Google Scholar] [CrossRef]
- Shimabukuro, R.H.; Hoffer, B.L. Metabolism of diclofop methyl in susceptible and resistant biotypes of Lolium rigidum. Pestic. Biochem. Physiol. 1991, 39, 251–260. [Google Scholar] [CrossRef]
- De Prado, R.; Franco, A.R. Cross-resistance and herbicide metabolism in grass weeds in Europe: Biochemical and physiological aspects. Weed Sci. 2004, 52, 115–120. [Google Scholar] [CrossRef]
- Buhler, D.D.; Swisher, B.A.; Burnside, O.C. Basis for antagonism in mixtures of bromoxynil plus quizalofop-p applied to yellow foxtail. Weed Sci. 1985, 33, 291–299. [Google Scholar]
- Liebl, R.; Worsham, A.D. Effect of chlorsulfuron on the movement and fate of diclofop in Italian ryegrass (Lolium multiflorum) and wheat (Triticum aestivum). Weed Sci. 1987, 35, 623–628. [Google Scholar]
- Yu, Q.; Powles, S.B. Metabolism-based herbicide resistance and cross-resistance in crop weeds: A threat to herbicide sustainability and global crop production. Plant Physiol. 2014, 166, 1106–1118. [Google Scholar] [CrossRef] [PubMed]
- Martins, B.A.B.; Sánchez-Olguín, E.; Perez-Jones, A.; Hulting, A.G.; Mallory-Smith, C. Alleles contributing to ACCase-resistance in an Italian ryegrass (Lolium perenne ssp. multiflorum) population from Oregon. Weed Sci. 2014, 62, 468–473. [Google Scholar] [CrossRef]
- Kuk, Y.I.; Wu, J.R.; Derr, J.F.; Hatzios, K.K. Mechanism of fenoxaprop resistance in an accession of smooth crabgrass (Digitaria ischaemum). Pestic. Biochem. Phys. 1999, 64, 112–123. [Google Scholar] [CrossRef]
- Jang, S.; Marjanovic, J.; Gornicki, P. Resistance to herbicides caused by single amino acid mutations in acetyl-CoA carboxylase in resistant populations of grassy weeds. New Phytol. 2013, 197, 1110–1116. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Hipólito, H.; Osuna, M.D.; Heredia, A.; Ruiz-Santaella, J.P.; De Prado, R. Non target mechanism involved in glyphosate tolerance found in Canavalia ensiformis plants. J. Agric. Food Chem. 2009, 57, 4844–4848. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz-Hipolito, H.; Fernandez, P.; Alcantara, R.; Gherekhloo, J.; Osuna, M.D.; De Prado, R. Ile-1781-Leu and Asp-2078-Gly Mutations in ACCase Gene, Endow Cross-resistance to APP, CHD, and PPZ in Phalaris minor from Mexico. Int. J. Mol. Sci. 2015, 16, 21363-21377. https://doi.org/10.3390/ijms160921363
Cruz-Hipolito H, Fernandez P, Alcantara R, Gherekhloo J, Osuna MD, De Prado R. Ile-1781-Leu and Asp-2078-Gly Mutations in ACCase Gene, Endow Cross-resistance to APP, CHD, and PPZ in Phalaris minor from Mexico. International Journal of Molecular Sciences. 2015; 16(9):21363-21377. https://doi.org/10.3390/ijms160921363
Chicago/Turabian StyleCruz-Hipolito, Hugo, Pablo Fernandez, Ricardo Alcantara, Javid Gherekhloo, Maria Dolores Osuna, and Rafael De Prado. 2015. "Ile-1781-Leu and Asp-2078-Gly Mutations in ACCase Gene, Endow Cross-resistance to APP, CHD, and PPZ in Phalaris minor from Mexico" International Journal of Molecular Sciences 16, no. 9: 21363-21377. https://doi.org/10.3390/ijms160921363







