Exploring the Molecular Interactions of 7,8-Dihydroxyflavone and Its Derivatives with TrkB and VEGFR2 Proteins
Abstract
:1. Introduction
2. Results
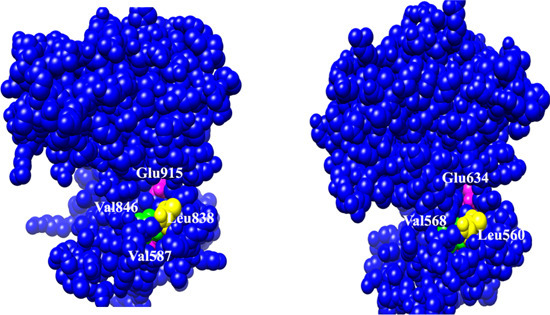
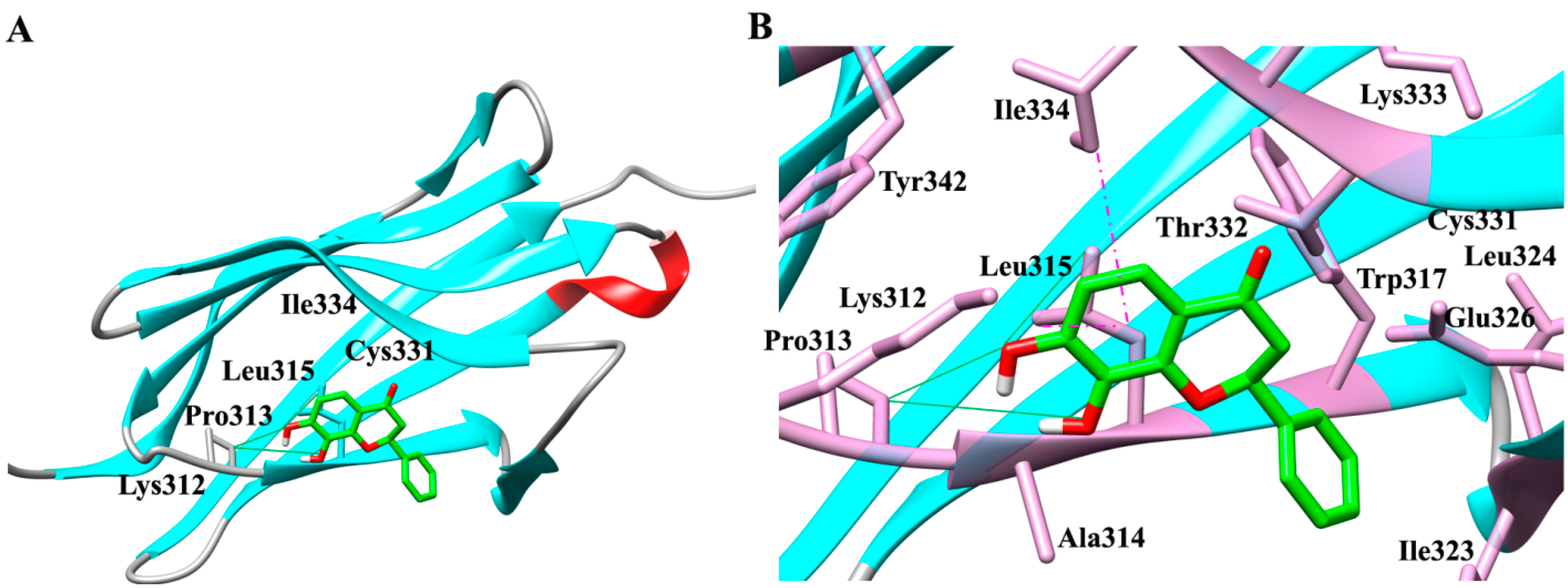
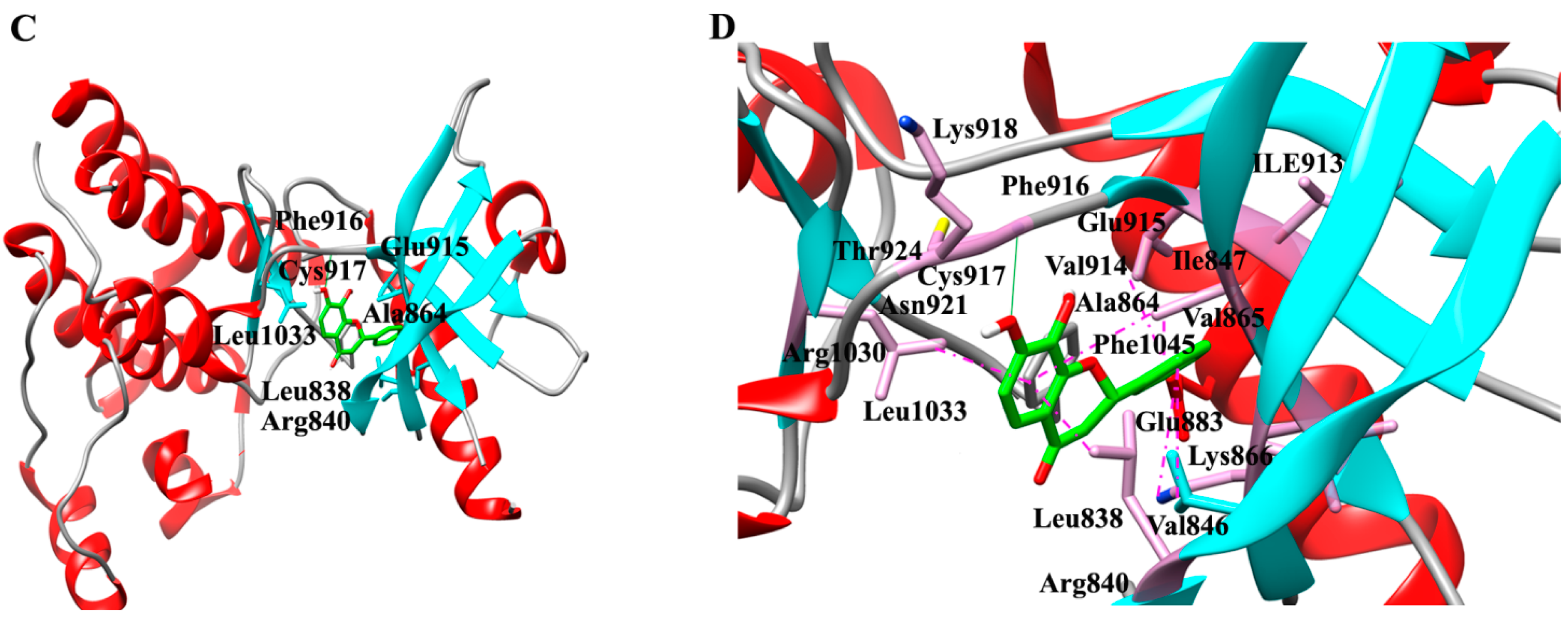
2.1. Molecular Determinants of 7,8-DHF Binding with TrkB and VEGFR2
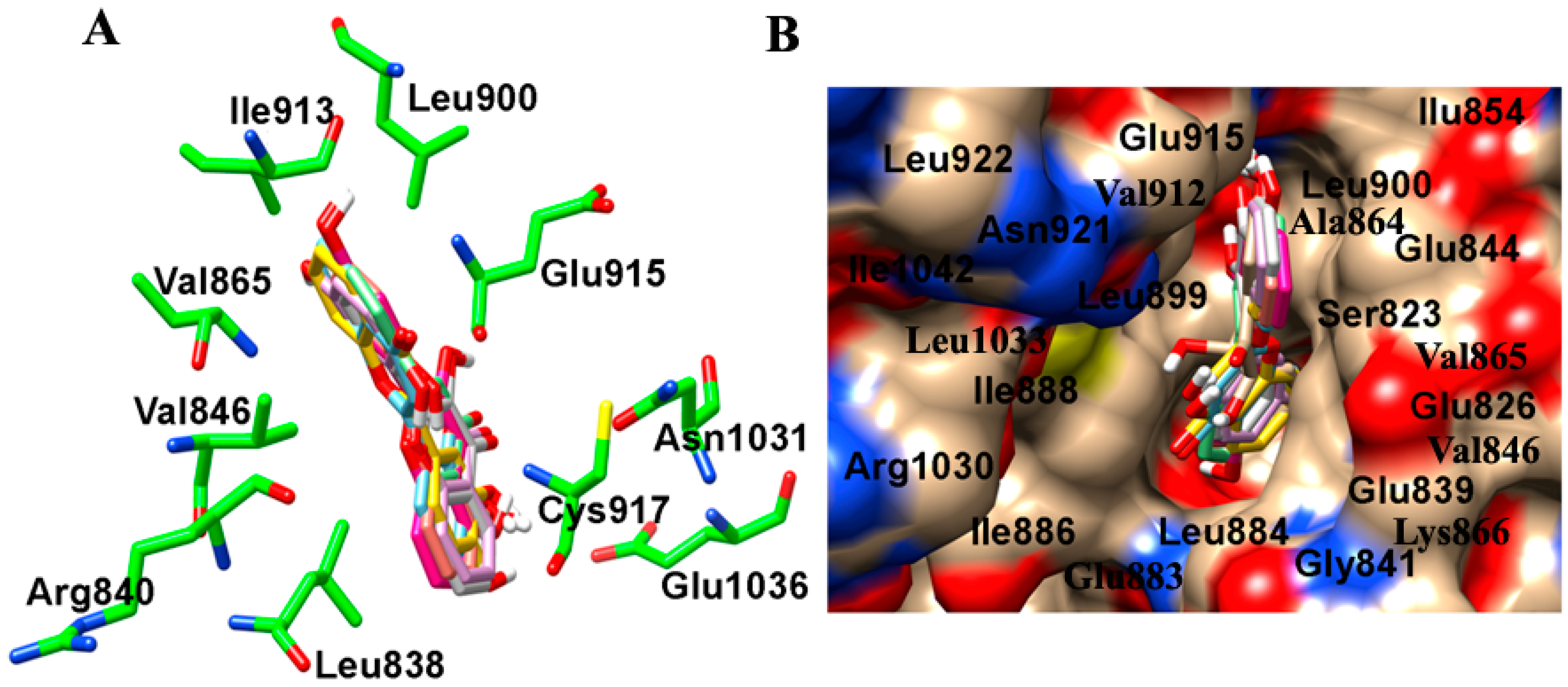
2.2. Binding Interactions of TrkB and VEGFR2 Receptors with 7,8-DHF Derivatives
| Compounds | Ring A | Ring B | Ring C | Scaffolds |
|---|---|---|---|---|
| 7,8-DHF | 7,8 di-OH | – | – | I |
| C1 | – | 2,3 di-OH | – | I |
| C2 | – | – | 2ʹ,3ʹ di-OH | II |
| C3 | – | – | 2ʹ,5ʹ di-OH | II |
| C4 | 5-OH | – | 2ʹ-OH | I and II |
| C5 | – | – | 2ʹ,5ʹ di-OH, 5ʹ-acetate | II |
| C6 | 5-OH | – | 4ʹ-OH | I and II |
| C7 | – | 3-OH | 2ʹ-OH | I and II |
| C8 | – | 3-OH | 4ʹ-OH | I and II |
| C9 | – | – | 3ʹ,4ʹ di-OH | II |
| C10 | – | – | 3ʹ,4ʹ di-OH, 4ʹ-glucoside | II |
| C11 | 5-OH, 6,7 di-methoxy | 3-methoxy | 3ʹ-OH, 4ʹ-methoxy | I and II |
| C12 | 3,5 di-OH | – | – | I |
| C13 | – | – | 3ʹ,5ʹ di-OH | II |
| C14 | 3,6 di-OH | – | – | I |
| C15 | 3,7 di-OH | – | – | I |
| C16 | 6-OH | – | 4ʹ-OH | I and II |
| C17 | 7-OH | – | 4ʹ-OH | I and II |
| C18 | 5-OH | – | 3ʹ-OH | I and II |
| C19 | 5-OH, 7-methoxy | – | 4ʹ-OH, 3ʹ-methoxy | I and II |
| C20 | 5,6 di-OH | – | – | I |
| C21 | 5,7 di-OH | – | 4ʹ-methoxy | I and II |
| C22 | 5,7 di-OH | – | – | I |
| C23 | 5,7 di-OH | – | 7-benzoate | I and II |
| C24 | 5,7 di-OH, 7 β-monoglucoside | – | – | I |
| C25 | 5,8 di-OH | – | – | I |
| C26 | 6,7 di-OH | – | – | I |
| C27 | 6,8 di-Cl | – | 3ʹ,5ʹ di-OH | I and II |
| C28 | 5-OH, 6-methoxy, 7-O-glucoside | – | 4ʹ-OH | I and II |
| C29 | 7-OH | – | 2ʹ-OH | I and II |
| C30 | 7-OH, 7-glucoside | – | 2ʹ-OH | I and II |
| C31 | 7-OH | – | 3ʹ-OH | I and II |
| C32 | 7-OH, 7-glucoside | – | 4ʹ-OH | I and II |
| C33 | 7-OH, 7-rutinoside | – | 4ʹ-OH | I and II |
| C34 | 5, 6-OH, 7-d-glucuronic acid | – | – | I |
| C35 | 8-OH | – | 2ʹ-OH | I and II |
| C36 | 7-OH, 8-β-d-glucopyranosyl | – | 4ʹ-OH | I and II |
| Compound Name | BEe (kcal/mol) | Ki (µM) | IMEe (kcal/mol) | Vdw–Hb–Ds (kcal/mol) | Ee (kcal/mol) | IEe (kcal/mol) | TFEe (kcal/mol) |
|---|---|---|---|---|---|---|---|
| 7,8-DHF | −5.71 | 64.79 | −5.84 | −5.6 | −0.24 | −0.69 | 0.82 |
| C1 | −5.94 | 44.35 | −5.97 | −5.91 | −0.06 | −0.8 | 0.82 |
| C2 | −5.91 | 46.34 | −6.57 | −6.12 | −0.45 | −0.16 | 0.82 |
| C3 | −5.28 | 133.89 | −5.54 | −5.34 | −0.2 | −0.57 | 0.82 |
| C4 | −6.13 | 32.06 | −6.28 | −5.94 | −0.34 | −0.67 | 0.82 |
| C5 | −5.96 | 43.12 | −6.84 | −6.41 | −0.43 | −0.21 | 1.10 |
| C6 | −5.65 | 71.59 | −5.77 | −5.63 | −0.13 | −0.71 | 0.82 |
| C7 | −5.63 | 75.12 | −6.09 | −5.71 | −0.38 | −0.36 | 0.82 |
| C8 | −5.52 | 89.37 | −5.95 | −5.85 | −0.09 | −0.4 | 0.82 |
| C9 | −6.02 | 38.62 | −6.7 | −6.51 | −0.19 | −0.14 | 0.82 |
| C10 | −7.10 | 6.21 | −6.33 | −6.08 | −0.25 | −3.24 | 2.47 |
| C11 | −7.42 | 3.62 | −7.67 | −7.49 | −0.17 | −1.68 | 1.92 |
| C12 | −6.43 | 19.41 | −6.36 | −6.01 | −0.35 | −0.89 | 0.82 |
| C13 | −5.18 | 158.66 | −5.85 | −5.6 | −0.25 | −0.15 | 0.82 |
| C14 | −5.24 | 143.46 | −5.51 | −5.37 | −0.14 | −0.56 | 0.82 |
| C15 | −5.36 | 118.3 | −5.86 | −5.58 | −0.28 | −0.32 | 0.82 |
| C16 | −5.28 | 135.23 | −5.94 | −5.76 | −0.18 | −0.16 | 0.82 |
| C17 | −5.49 | 94.66 | −6.14 | −5.97 | −0.18 | −0.17 | 0.82 |
| C18 | −5.81 | 55.47 | −5.95 | −5.67 | −0.28 | −0.68 | 0.82 |
| C19 | −6.17 | 30.1 | −6.37 | −5.99 | −0.38 | −1.17 | 1.37 |
| C20 | −6.00 | 39.92 | −5.81 | −5.52 | −0.29 | −1.01 | 0.82 |
| C21 | −5.97 | 42.38 | −6.30 | −6.11 | −0.19 | −0.76 | 1.10 |
| C22 | −5.46 | 98.9 | −5.56 | −5.36 | −0.20 | −0.73 | 0.82 |
| C23 | −6.47 | 18.12 | −6.68 | −6.51 | −0.17 | −1.16 | 1.37 |
| C24 | −6.85 | 9.56 | −7.02 | −6.88 | −0.13 | −2.3 | 2.47 |
| C25 | −6.08 | 35.18 | −5.82 | −5.78 | −0.04 | −1.08 | 0.82 |
| C27 | −6.29 | 24.48 | −6.92 | −6.72 | −0.2 | −0.19 | 0.82 |
| C28 | −7.90 | 1.61 | −8.31 | −8.05 | −0.27 | −2.61 | 3.02 |
| C29 | −5.41 | 108.5 | −6.05 | −5.76 | −0.29 | −0.18 | 0.82 |
| C30 | −6.53 | 16.21 | −7.06 | −6.84 | −0.22 | −1.94 | 2.47 |
| C31 | −5.62 | 76.09 | −6.29 | −5.91 | −0.38 | −0.15 | 0.82 |
| C32 | −5.89 | 47.88 | −6.25 | −5.99 | −0.26 | −2.11 | 2.47 |
| C33 | −6.16 | 4.82 | −4.05 | −3.86 | −0.19 | −2.68 | 3.57 |
| C34 | −7.50 | 3.18 | −6.97 | −6.38 | −0.6 | −3.27 | 2.74 |
| C35 | −5.64 | 72.83 | −5.68 | −5.26 | −0.42 | −0.79 | 0.82 |
| C36 | −6.77 | 10.99 | −7.78 | −7.15 | −0.63 | −1.45 | 2.47 |
| Compound Name | BEe (kcal/mol) | Ki (µM) | IMEe (kcal/mol) | Vdw-Hb-Ds (kcal/mol) | Ee (kcal/mol) | IEe (kcal/mol) | TFEe (kcal/mol) |
|---|---|---|---|---|---|---|---|
| AAX | −9.68 | 0.08 | −10.43 | −10.42 | −0.01 | −1.44 | +2.20 |
| 7,8-DHF | −7.76 | 2.04 | −8.09 | −7.97 | −0.12 | −0.50 | +0.82 |
| C1 | −6.69 | 12.49 | −6.49 | −6.49 | +0.00 | −1.03 | +0.82 |
| C2 | −7.01 | 7.29 | −7.28 | −7.18 | −0.10 | −0.56 | +0.82 |
| C3 | −6.66 | 13.11 | −6.91 | −6.86 | −0.05 | −0.57 | +0.82 |
| C4 | −6.97 | 7.72 | −7.17 | −7.09 | −0.08 | −0.62 | +0.82 |
| C5 | −7.44 | 3.51 | −8.27 | −8.08 | −0.19 | −0.27 | +1.10 |
| C6 | −7.27 | 4.66 | −7.39 | −7.23 | −0.15 | −0.71 | +0.82 |
| C7 | −7.08 | 6.44 | −7.09 | −6.97 | −0.11 | −0.82 | +0.82 |
| C8 | −7.19 | 5.41 | −7.52 | −7.25 | −0.27 | −0.49 | +0.82 |
| C9 | −7.07 | 6.52 | −7.76 | −7.59 | −0.17 | −0.14 | +0.82 |
| C10 | −9.41 | 0.13 | −9.64 | −9.20 | −0.44 | −2.24 | +2.47 |
| C11 | −6.33 | 22.82 | −6.69 | −6.49 | −0.20 | −1.56 | +1.92 |
| C12 | −7.41 | 3.71 | −7.16 | −7.13 | −0.03 | −1.07 | +0.82 |
| C13 | −7.25 | 4.84 | −7.91 | −7.58 | −0.33 | −0.17 | +0.82 |
| C14 | −7.31 | 4.41 | −7.61 | −7.48 | −0.14 | −0.52 | +0.82 |
| C15 | −7.30 | 4.46 | −7.69 | −7.56 | −0.13 | −0.44 | +0.82 |
| C16 | −7.27 | 4.69 | −7.93 | −7.68 | −0.25 | −0.17 | +0.82 |
| C17 | −7.54 | 2.99 | −8.19 | −7.89 | −0.30 | −0.17 | +0.82 |
| C18 | −7.08 | 6.48 | −7.19 | −6.88 | −0.31 | −0.71 | +0.82 |
| C19 | −7.73 | 2.16 | −7.92 | −7.79 | −0.13 | −1.18 | +1.37 |
| C20 | −7.75 | 2.10 | −7.87 | −7.73 | −0.14 | −0.70 | +0.82 |
| C21 | −7.23 | 5.03 | −7.56 | −7.48 | −0.08 | −0.77 | +1.10 |
| C22 | −7.53 | 3.02 | −7.61 | −7.55 | −0.06 | −0.74 | +0.82 |
| C23 | −8.34 | 0.77 | −8.61 | −8.56 | −0.05 | −1.10 | +1.37 |
| C24 | −8.17 | 1.03 | −8.47 | −8.23 | −0.24 | −2.17 | +2.47 |
| C25 | −7.17 | 5.54 | −7.04 | −6.99 | −0.05 | −0.95 | +0.82 |
| C26 | −7.60 | 2.68 | −7.77 | −7.58 | −0.19 | −0.65 | +0.82 |
| C27 | −7.50 | 3.17 | −8.14 | −7.85 | −0.29 | −0.19 | +0.82 |
| C28 | −8.76 | 0.38 | −9.01 | −8.76 | −0.25 | −2.77 | +3.02 |
| C29 | −6.93 | 8.28 | −7.16 | −7.12 | −0.04 | −0.59 | +0.82 |
| C30 | −7.96 | 1.47 | −8.23 | −7.81 | −0.42 | −2.20 | +2.47 |
| C31 | −7.46 | 3.40 | −8.11 | −7.65 | −0.47 | −0.17 | +0.82 |
| C32 | −8.43 | 0.66 | −9.03 | −8.68 | −0.36 | −1.86 | +2.47 |
| C33 | −8.14 | 1.09 | −7.83 | −7.65 | −0.18 | −3.88 | +3.57 |
| C34 | −9.22 | 0.174 | −9.45 | −8.47 | −0.98 | −2.52 | +2.74 |
| C35 | −6.73 | 11.57 | −6.60 | −6.56 | −0.04 | −0.96 | +0.82 |
| C36 | −8.29 | 0.84 | −8.21 | −8.07 | −0.14 | −2.55 | +2.47 |

2.3. Molecular Dynamics (MD) of 7,8-DHF-TrkB and 7,8-DHF-VEGFR2 Complex


2.4. 7,8-DHF Treatment Leads to Loss of VEGFR2 Activity
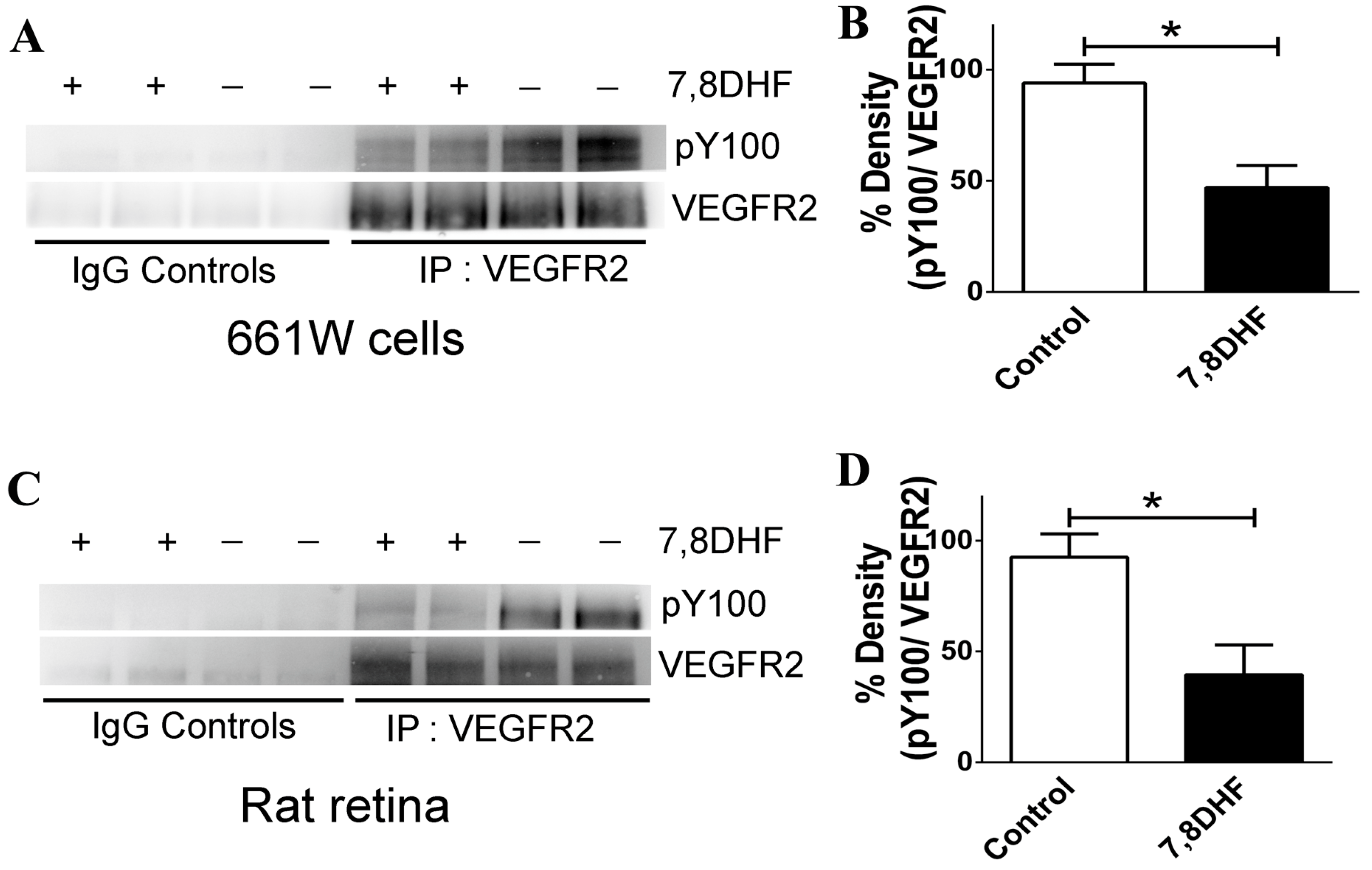
3. Discussion
4. Experimental Section
4.1. Chemicals
4.2. Animal Experiments
4.3. Selection and Preparation of Macromolecule
4.4. Selection and Preparation of Dihydroxy Flavones Derivatives
4.5. Molecular Docking
4.6. Molecular Dynamics Simulations
4.7. Cell Culture and Treatment Regimens
4.8. Western Blot and Immunoprecipitations
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jang, S.W.; Liu, X.; Yepes, M.; Shepherd, K.R.; Miller, G.W.; Liu, Y.; Wilson, W.D.; Xiao, G.; Blanchi, B.; Sun, Y.E.; et al. A selective TrkB agonist with potent neurotrophic activities by 7,8-dihydroxyflavone. Proc. Natl. Acad. Sci. USA 2010, 107, 2687–2692. [Google Scholar] [CrossRef] [PubMed]
- Castello, N.A.; Nguyen, M.H.; Tran, J.D.; Cheng, D.; Green, K.N.; LaFerla, F.M. 7,8-Dihydroxyflavone, a small molecule TrkB agonist, improves spatial memory and increases thin spine density in a mouse model of alzheimer disease-like neuronal loss. PLoS ONE 2014, 9, e91453. [Google Scholar] [CrossRef] [PubMed]
- Chapleau, C.A.; Lane, J.; Larimore, J.; Li, W.; Pozzo-Miller, L.; Percy, A.K. Recent progress in Rett syndrome and MECP2 dysfunction: Assessment of potential treatment options. Future Neurol. 2013, 8, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Peng, Q.; Liu, X.; Jin, J.; Hou, Z.; Zhang, J.; Mori, S.; Ross, C.A.; Ye, K.; Duan, W. Small-molecule TrkB receptor agonists improve motor function and extend survival in a mouse model of huntington’s disease. Hum. Mol. Genet. 2013, 22, 2462–2470. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chan, C.B.; Jang, S.W.; Pradoldej, S.; Huang, J.; He, K.; Phun, L.H.; France, S.; Xiao, G.; Jia, Y.; et al. A synthetic 7,8-dihydroxyflavone derivative promotes neurogenesis and exhibits potent antidepressant effect. J. Med. Chem. 2010, 53, 8274–8286. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; You, Y.; Li, J.C.; Klistorner, A.; Graham, S.L. Protective effects of 7,8-dihydroxyflavone on retinal ganglion and RGC-5 cells against excitotoxic and oxidative stress. J. Mol. Neurosci. 2013, 49, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; You, Y.; Klistorner, A.; Graham, S.L. Shp-2 regulates the TrkB receptor activity in the retinal ganglion cells under glaucomatous stress. Biochim. Biophys. Acta. 2012, 1822, 1643–1649. [Google Scholar] [CrossRef] [PubMed]
- Haniu, M.; Talvenheimo, J.; Le, J.; Katta, V.; Welcher, A.; Rohde, M.F. Extracellular domain of neurotrophin receptor TrkB: Disulfide structure, N-glycosylation sites, and ligand binding. Arch. Biochem. Biophys. 1995, 322, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, M. Vascular endothelial growth factor (VEGF)-receptor2: Its biological functions, major signaling pathway, and specific ligand VEGF-E. Endothelium 2006, 13, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Hagstrom, S.A.; Ying, G.S.; Pauer, G.J.; Sturgill-Short, G.M.; Huang, J.; Maguire, M.G.; Martin, D.F. VEGFA and VEGFR2 gene polymorphisms and response to anti-vascular endothelial growth factor therapy: Comparison of age-related macular degeneration treatments trials (CATT). JAMA Ophthalmol. 2014, 132, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Daneshvar, R. Anti-VEGF agents and glaucoma filtering surgery. J. Ophthalmic Vis. Res. 2013, 8, 182–186. [Google Scholar] [PubMed]
- Holmes, K.; Roberts, O.L.; Thomas, A.M.; Cross, M.J. Vascular endothelial growth factor receptor-2: Structure, function, intracellular signalling and therapeutic inhibition. Cell. Signal. 2007, 19, 2003–2012. [Google Scholar] [CrossRef] [PubMed]
- Nishiguchi, K.M.; Nakamura, M.; Kaneko, H.; Kachi, S.; Terasaki, H. The role of VEGF and VEGFR2/Flk1 in proliferation of retinal progenitor cells in murine retinal degeneration. Investig. Ophthalmol. Vis. Sci. 2007, 48, 4315–4320. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Wu, Y.; Lin, L.; Wang, J.; Chen, Y.; Yi, Z.; Liu, M.; Pang, X. Hispidulin, a small flavonoid molecule, suppresses the angiogenesis and growth of human pancreatic cancer by targeting vascular endothelial growth factor receptor 2-mediated PI3K/Akt/mTOR signaling pathway. Cancer Sci. 2011, 102, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, N.; Han, S.; Wang, D.; Mo, S.; Yu, L.; Huang, H.; Tsui, K.; Shen, J.; Chen, J. Dietary compound isoliquiritigenin inhibits breast cancer neoangiogenesis via VEGF/VEGFR-2 signaling pathway. PLoS ONE 2013, 8, e68566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardy, B.; Douglas, N.; Helma, C.; Rautenberg, M.; Jeliazkova, N.; Jeliazkov, V.; Nikolova, I.; Benigni, R.; Tcheremenskaia, O.; Kramer, S.; et al. Collaborative development of predictive toxicology applications. J. Cheminform. 2010, 2, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, Y.; Kim, E.T.; Eastwood, M.P.; Dror, R.O.; Seeliger, M.A.; Shaw, D.E. How does a drug molecule find its target binding site? J. Am. Chem. Soc. 2011, 133, 9181–9183. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Lv, F.; Li, L.; Yu, H.; Dong, M.; Fu, Q. 7,8-dihydroxyflavone rescues spatial memory and synaptic plasticity in cognitively impaired aged rats. J. Neurochem. 2012, 122, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; You, Y.; Gupta, V.B.; Klistorner, A.; Graham, S.L. TrkB receptor signalling: Implications in neurodegenerative, psychiatric and proliferative disorders. Int. J. Mol. Sci. 2013, 14, 10122–10142. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H. Flavonoids inhibit VEGF/BFGF-induced angiogenesis in vitro by inhibiting the matrix-degrading proteases. J. Cell. Biochem. 2003, 89, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Le Bail, J.C.; Varnat, F.; Nicolas, J.C.; Habrioux, G. Estrogenic and antiproliferative activities on MCF-7 human breast cancer cells by flavonoids. Cancer Lett. 1998, 130, 209–216. [Google Scholar] [CrossRef]
- Lin, C.; Wu, M.; Dong, J. Quercetin-4ʹ-O-β-d-glucopyranoside (QODG) inhibits angiogenesis by suppressing VEGFR2-mediated signaling in zebrafish and endothelial cells. PLoS ONE 2012, 7, e31708. [Google Scholar] [CrossRef] [PubMed]
- Paramashivam, S.K.; Elayaperumal, K.; Natarajan, B.B.; Ramamoorthy, M.D.; Balasubramanian, S.; Dhiraviam, K.N. In silico pharmacokinetic and molecular docking studies of small molecules derived from indigofera aspalathoides vahl targeting receptor tyrosine kinases. Bioinformation 2015, 11, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Gowda, L.R. α-1-proteinase inhibitor is a heparin binding serpin: Molecular interactions with the lys rich cluster of helix-f domain. Biochimie 2008, 90, 749–761. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.J.; Vajda, S. Protein docking along smooth association pathways. Proc. Natl. Acad. Sci. USA 2001, 98, 10636–10641. [Google Scholar] [CrossRef] [PubMed]
- Tsui, L.; Fong, T.H.; Wang, I.J. The effect of 3-(5ʹ-hydroxymethyl-2ʹ-furyl)-1-benzylindazole (YC-1) on cell viability under hypoxia. Mol. Vis. 2013, 19, 2260–2273. [Google Scholar] [PubMed]
- Meyer, M.; Clauss, M.; Lepple-Wienhues, A.; Waltenberger, J.; Augustin, H.G.; Ziche, M.; Lanz, C.; Buttner, M.; Rziha, H.J.; Dehio, C. A novel vascular endothelial growth factor encoded by orf virus, VEGF-E, mediates angiogenesis via signalling through VEGFR-2 (KDR) but not VEGFR-1 (Flt-1) receptor tyrosine kinases. EMBO J. 1999, 18, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Kroll, J.; Waltenberger, J. The vascular endothelial growth factor receptor KDR activates multiple signal transduction pathways in porcine aortic endothelial cells. J. Biol. Chem. 1997, 272, 32521–32527. [Google Scholar] [CrossRef] [PubMed]
- Karkkainen, M.J.; Petrova, T.V. Vascular endothelial growth factor receptors in the regulation of angiogenesis and lymphangiogenesis. Oncogene 2000, 19, 5598–5605. [Google Scholar] [CrossRef] [PubMed]
- Claesson-Welsh, L. Signal transduction by vascular endothelial growth factor receptors. Biochem. Soc. Trans. 2003, 31, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Banfield, M.J.; Naylor, R.L.; Robertson, A.G.; Allen, S.J.; Dawbarn, D.; Brady, R.L. Specificity in Trk receptor:Neurotrophin interactions: The crystal structure of TrkB-D5 in complex with neurotrophin-4/5. Structure 2001, 9, 1191–1199. [Google Scholar] [CrossRef]
- Harris, P.A.; Cheung, M.; Hunter, R.N., 3rd.; Brown, M.L.; Veal, J.M.; Nolte, R.T.; Wang, L.; Liu, W.; Crosby, R.M.; Johnson, J.H.; et al. Discovery and evaluation of 2-anilino-5-aryloxazoles as a novel class of VEGFR2 kinase inhibitors. J. Med. Chem. 2005, 48, 1610–1619. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide protein data bank. Nat. Struct. Biol. 2003, 10, 980. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. Ucsf chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Dundas, J.; Ouyang, Z.; Tseng, J.; Binkowski, A.; Turpaz, Y.; Liang, J. CASTp: Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006, 34, W116–W118. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Barrett, T.; Benson, D.A.; Bolton, E.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; di Cuccio, M.; Federhen, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2011, 39, D38–D51. [Google Scholar] [CrossRef] [PubMed]
- Chitranshi, N.; Gupta, S.; Tripathi, P.K.; Seth, P.K. New molecular scaffolds for the design of alzheimer’s acetylcholinesterase inhibitors identified using ligand- and receptor-based virtual screening. Med. Chem. Res. 2013, 22, 2328–2345. [Google Scholar] [CrossRef]
- Prasanna, S.; Manivannan, E.; Chaturvedi, S.C. Quantitative structure–activity relationship analysis of 2,3-diaryl indoles as selective cyclooxygenase-2 inhibitors. J. Enzyme Inhib. Med. Chem. 2005, 20, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and autodocktools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Bikadi, Z.; Hazai, E. Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of autodock. J. Cheminform. 2009, 1, 15. [Google Scholar] [CrossRef] [PubMed]
- Labute, P. Protonate3d: Assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins 2009, 75, 187–205. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, C. Multiple ligand simultaneous docking: Orchestrated dancing of ligands in binding sites of protein. J. Comput. Chem. 2010, 31, 2014–2022. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.E.; Brown, D.G.; Mills, J.E.; Phillips, C.; Morris, G. Computational tools for the analysis and visualization of multiple protein–ligand complexes. J. Mol. Graph. Model. 2005, 24, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. Ligplot+: Multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Mohanty, U.; Noehre, J.; Sawyer, T.K.; Sherman, W.; Krilov, G. Probing the α-helical structural stability of stapled p53 peptides: Molecular dynamics simulations and analysis. Chem. Biol. Drug Des. 2010, 75, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Siepmann, J.I.; Schure, M.R. Conformation and solvation structure for an isolated n-octadecane chain in water, methanol, and their mixtures. J. Phys. Chem. B 2006, 110, 10519–10525. [Google Scholar] [CrossRef] [PubMed]
- Cerutti, D.S.; Duke, R.E.; Darden, T.A.; Lybrand, T.P. Staggered mesh Ewald: An extension of the smooth particle-mesh ewald method adding great versatility. J. Chem. Theory Comput. 2009, 5, 2322. [Google Scholar] [CrossRef] [PubMed]
- Strahan, G.D.; Keniry, M.A.; Shafer, R.H. NMR structure refinement and dynamics of the K+-[D(G3T4G3)]2 quadruplex via particle mesh ewald molecular dynamics simulations. Biophys. J. 1998, 75, 968–981. [Google Scholar] [CrossRef]
- Basavarajappa, D.K.; Gupta, V.K.; Dighe, R.; Rajala, A.; Rajala, R.V. Phosphorylated GRB14 is an endogenous inhibitor of retinal protein tyrosine phosphatase 1b, and light-dependent activation of Src phosphorylates GRB14. Mol. Cell. Biol. 2011, 31, 3975–3987. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Rajala, A.; Rajala, R.V. Insulin receptor regulates photoreceptor cng channel activity. Am. J. Physiol. Endocrinol. Metab. 2012, 303, E1363–E1372. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; You, Y.; Li, J.; Gupta, V.; Golzan, M.; Klistorner, A.; van den Buuse, M.; Graham, S. BDNF impairment is associated with age-related changes in the inner retina and exacerbates experimental glaucoma. Biochim. Biophys. Acta 2014, 1842, 1567–1578. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Rajala, A.; Daly, R.J.; Rajala, R.V. Growth factor receptor-bound protein 14: A new modulator of photoreceptor-specific cyclic-nucleotide-gated channel. EMBO Rep. 2010, 11, 861–867. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chitranshi, N.; Gupta, V.; Kumar, S.; Graham, S.L. Exploring the Molecular Interactions of 7,8-Dihydroxyflavone and Its Derivatives with TrkB and VEGFR2 Proteins. Int. J. Mol. Sci. 2015, 16, 21087-21108. https://doi.org/10.3390/ijms160921087
Chitranshi N, Gupta V, Kumar S, Graham SL. Exploring the Molecular Interactions of 7,8-Dihydroxyflavone and Its Derivatives with TrkB and VEGFR2 Proteins. International Journal of Molecular Sciences. 2015; 16(9):21087-21108. https://doi.org/10.3390/ijms160921087
Chicago/Turabian StyleChitranshi, Nitin, Vivek Gupta, Sanjay Kumar, and Stuart L. Graham. 2015. "Exploring the Molecular Interactions of 7,8-Dihydroxyflavone and Its Derivatives with TrkB and VEGFR2 Proteins" International Journal of Molecular Sciences 16, no. 9: 21087-21108. https://doi.org/10.3390/ijms160921087