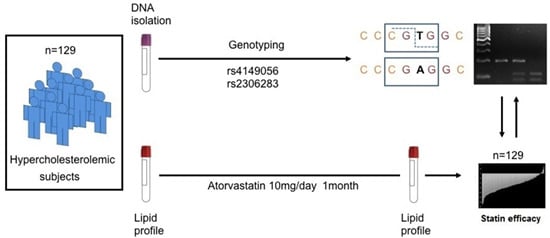
SLCO1B1 c.388A>G Polymorphism Is Associated with HDL-C Levels in Response to Atorvastatin in Chilean Individuals
Abstract
:1. Introduction
2. Results
2.1. Demographics and Clinical Variables
| Parameter | n = 129 |
|---|---|
| Age (years) | 56.2 ± 10.9 |
| Men/women (n) | 49/80 |
| BMI (kg/m2) | 25.6 ± 2.7 |
| Systolic blood pressure (mmHg) | 106.8 ± 12.3 |
| Diastolic blood pressure (mmHg) | 72.7 ± 9.2 |
| Glucose (mg/dL) | 97.2 ± 9.6 |
| TC (mg/dL) | 274.9 ± 18.5 |
| TG (mg/dL) | 214.1 ± 51.9 |
| LDL-C (mg/dL) | 186.3 ± 17.4 |
| HDL-C (mg/dL) | 45.8 ± 8.3 |
| AST/GOT (U/L) | 24.4 ± 7.2 |
| ALT/GPT (U/L) | 22.7 ± 8.3 |
| CK (U/L) | 103.5 ± 77.5 |
| Lipids | Baseline (mg/dL) | Treatment (mg/dL) | Change (mg/dL) | p-Value |
|---|---|---|---|---|
| TC | 274.4 ± 18.3 | 224.5 ± 26.2 | −49.8 ± 30.0 | <0.001 |
| HDL-C | 46.4 ± 8.8 | 54.1 ± 6.7 | 7.6 ± 6.7 | <0.001 |
| LDL-C | 185.4 ± 17.5 | 137.3 ± 26.1 | −48.1 ± 31.6 | <0.001 |
| TG | 212.8 ± 50.5 | 165.9 ± 48.4 | −47.1 ± 43.2 | <0.001 |
2.2. SLCO1B1 Single Nucleotide Polymorphisms
| SNP | Genotypes | Alleles | |||
|---|---|---|---|---|---|
| rs2306283 (388A>G) | AA | AG | GG | A 0.453 | G 0.547 |
| 18.6% | 53.5% | 27.9% | |||
| (n = 24) | (n = 69) | (n = 36) | |||
| rs4149056 (521T>C) | TT | TC | CC | T 0.864 | C 0.136 |
| 73.6% | 25.6% | 0.8% | |||
| (n = 95) | (n = 33) | (n = 1) | |||
2.3. SLCO1B1 Polymorphisms and Atorvastatin Treatment
| Lipids (mg/dL) | Condition | Genotypes | p-Value | ||
|---|---|---|---|---|---|
| AA (n = 24) | AG (n = 69) | GG (n = 36) | |||
| TC | Basal | 275. 5 ± 20.7 | 275.4 ± 17.9 | 273.4 ± 18.5 | 0.81 |
| Treatment | 228.0 ± 30.3 | 224.5 ± 25.9 | 221.6 ± 26.4 | ||
| % Change | −17.0 ± 10.4 | −18.3 ± 10.8 | −18.9 ± 10.3 | ||
| HDL-C | Basal | 48.9 ± 10.8 | 45.1 ± 7.6 | 44.8 ± 7.3 | 0.02 * |
| Treatment | 53.4 ± 6.7 | 53.6 ± 7.6 | 54.0 ± 4.9 | ||
| % Change | 11.6 ± 14.7 a | 20.0 ± 13.9 b | 22.5 ± 16.6 b | ||
| LDL-C | Basal | 183.6 ± 21.2 | 187.3 ± 16.7 | 186.4 ± 15.9 | 0.27 |
| Treatment | 139.6 ± 28.8 | 138.5 ± 26.8 | 134.1 ± 25.5 | ||
| % Change | −21.6 ± 14.9 | −25.3 ± 15.9 | −28.3 ± 14.3 | ||
| TG | Basal | 216.7 ± 42.5 | 214.8 ± 50.8 | 210.9 ± 60.2 | 0.30 |
| Treatment | 174.2 ± 42.0 | 159.5 ± 49.6 | 164.0 ± 48.3 | ||
| % Change | −19.3 ± 15.4 | −24.9 ± 19.6 | −19.8 ± 20.5 | ||
| Lipids (mg/dL) | Condition | Genotypes | p-Value | |
|---|---|---|---|---|
| TT (n = 95) | TC + CC (n = 34) | |||
| TC | Basal | 273.6 ± 19.5 | 276.7 ± 16.0 | 0.58 |
| Treatment | 223.6 ± 27.0 | 223.0 ± 24.9 | ||
| % Change | −17.9 ± 10.8 | −19.1 ± 10.7 | ||
| HDL-C | Basal | 46.2 ± 9.2 | 44.3 ± 4.7 | 0.15 |
| Treatment | 53.5 ± 6.6 | 54.0 ± 6.2 | ||
| % Change | 17.9 ± 16.1 | 22.4 ± 13.2 | ||
| LDL-C | Basal | 185.1 ± 18.9 | 188.4 ± 12.8 | 0.34 |
| Treatment | 137.5 ± 26.7 | 135.0 ± 26.0 | ||
| % Change | −24.8 ± 16.4 | −27.9 ± 14.9 | ||
| TG | Basal | 211.8 ± 49.9 | 219.8 ± 57.0 | 0.92 |
| Treatment | 162.9 ± 46.0 | 169.9 ± 51.1 | ||
| % Change | −21.9 ± 18.5 | −21.5 ± 20.5 | ||
3. Discussion
4. Experimental Section
4.1. Subjects
4.2. Molecular Analysis
4.3. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Brown, M.S.; Goldstein, J.L. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 1997, 89, 331–340. [Google Scholar] [CrossRef]
- Wierzbicki, A.S.; Poston, R.; Ferro, A. The lipid and non-lipid effects of statins. Pharmacol. Ther. 2003, 99, 95–112. [Google Scholar] [CrossRef]
- Vaughan, C.J.; Gotto, A.M., Jr.; Basson, C.T. The evolving role of statins in the management of atherosclerosis. J. Am. Coll. Cardiol. 2000, 35, 1–10. [Google Scholar] [CrossRef]
- Naci, H.; Brugts, J.; Ades, T. Comparative tolerability and harms of individual statins: a study-level network meta-analysis of 246 955 participants from 135 randomized, controlled trials. Circ. Cardiovasc. Qual. Outcomes 2013, 6, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Stewart, A. SLCO1B1 polymorphisms and statin-induced myopathy. PLoS Curr. 2013. [Google Scholar] [CrossRef] [PubMed]
- Link, E.; Parish, S.; Armitage, J.; Bowman, L.; Heath, S.; Matsuda, F.; Gut, I.; Lathrop, M.; Collins, R. SLCO1B1 variants and statin-induced myopathy—A genomewide study. N. Engl. J. Med. 2008, 359, 789–799. [Google Scholar] [PubMed]
- Dendramis, G. Interindividual differences in the response to statin therapy and gene polymorphisms related to myopathy during statin therapy. G. Ital. Cardiol. 2011, 12, 182–185. [Google Scholar]
- Ulvestad, M.; Skottheim, I.B.; Jakobsen, G.S.; Bremer, S.; Molden, E.; Asberg, A.; Hjelmesæth, J.; Andersson, T.B.; Sandbu, R.; Christensen, H. Impact of OATP1B1, MDR1, and CYP3A4 expression in liver and intestine on interpatient pharmacokinetic variability of atorvastatin in obese subjects. Clin. Pharmacol. Ther. 2013, 93, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Sortica, V.A.; Fiegenbaum, M.; Lima, L.O.; van der Sand, C.R.; van der Sand, L.C.; Ferreira, M.E.W.; Pires, R.C.; Hutz, M.H. SLCO1B1 gene variability influences lipid-lowering efficacy on simvastatin therapy in Southern Brazilians. Clin. Chem. Lab. Med. 2012, 50, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Romaine, S.P.; Bailey, K.M.; Hall, A.S.; Balmforth, A.J. The influence of SLCO1B1 (OATP1B1) gene polymorphisms on response to statin therapy. Pharmacogenom. J. 2010, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tirona, R.G.; Leake, B.F.; Merino, G.; Kim, R.B. Polymorphisms in OATP-C: Identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J. Biol. Chem. 2001, 276, 35669–35675. [Google Scholar] [CrossRef] [PubMed]
- De Keyser, C.E.; Peters, B.J.; Becker, M.L.; Visser, L.E.; Uitterlinden, A.G.; Klungel, O.H.; Verstuyft, C.; Hofman, A.; Maitland-van der Zee, A.H.; Stricker, B.H. The SLCO1B1 c.521T>C polymorphism is associated with dose decrease or switching during statin therapy in the Rotterdam Study. Pharmacogenet. Genom. 2014, 24, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Mwinyi, J.; Johne, A.; Bauer, S.; Roots, I.; Gerloff, T. Evidence for inverse effects of OATP-C (SLC21A6) 5 and 1b haplotypes on pravastatin kinetics. Clin. Pharmacol. Ther. 2004, 75, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Niemi, M.; Schaeffeler, E.; Lang, T.; Fromm, M.F.; Neuvonen, M.; Kyrklund, C.; Backman, J.T.; Kerb, R.; Schwab, M.; Neuvonen, P.; et al. High plasma pravastatin concentrations are associated with single nucleotide polymorphisms and haplotypes of organic anion transporting polypeptide-C (OATP-C, SLCO1B1). Pharmacogenetics 2004, 14, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Kalliokoski, A.; Neuvonen, P.J.; Niemi, M. SLCO1B1 polymorphism and oral antidiabetic drugs. Basic Clin. Pharmacol. Toxicol. 2010, 107, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Vergeer, M.; Holleboom, A.G.; Kastelein, J.J.; Kuivenhoven, J.A. The HDL hypothesis: Does high-density lipoprotein protect from atherosclerosis? J. Lipid Res. 2010, 51, 2058–2073. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.C.; Perin, P.M.; Purim, S.G.; Silbiger, V.N.; Genvigir, F.D.; Willrich, M.A.; Arazi, S.S.; Luchessi, A.D.; Hirata, M.H.; Bernik, M.M.S.; et al. Pharmacogenetics of OATP transporters reveals that SLCO1B1 c.388A>G variant is determinant of increased atorvastatin response. Int. J. Mol. Sci. 2011, 12, 5815–5827. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Li, Y.P.; Gao, Y.; Yang, S.H.; Lu, P.Q.; Jia, M.; Zhang, L.-R. Lack of association between SLCO1B1 polymorphism and the lipid-lowering effects of atorvastatin and simvastatin in Chinese individuals. Eur. J. Clin. Pharmacol. 2013, 69, 1269–1274. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chen, B.L.; Ozdemir, V.; He, Y.J.; Zhou, G.; Peng, D.D.; Deng, S.; Xie, Q.Y.; Xie, W.; Xu, L.Y.; et al. SLCO1B1 521T→C functional genetic polymorphism and lipid-lowering efficacy of multiple-dose pravastatin in Chinese coronary heart disease patients. Br. J. Clin. Pharmacol. 2007, 64, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Rosales, A.; Alvear, M.; Cuevas, A.; Saavedra, N.; Zambrano, T.; Salazar, L.A. Identification of pharmacogenetic predictors of lipid-lowering response to atorvastatin in Chilean subjects with hypercholesterolemia. Clin. Chim. Acta 2012, 413, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Shabana, M.F.; Mishriki, A.A.; Issac, M.S.; Bakhoum, S.W. Do MDR1 and SLCO1B1 polymorphisms influence the therapeutic response to atorvastatin? A study on a cohort of Egyptian patients with hypercholesterolemia. Mol. Diagn. Ther. 2013, 17, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.H.; McKenney, J.M.; Karalis, D.G.; Downey, J. Comparison of the efficacy and safety of atorvastatin initiated at different starting doses in patients with dyslipidemia. Am. Heart J. 2005, 149, e1–e8. [Google Scholar] [CrossRef] [PubMed]
- Zambrano, T.; Hirata, M.H.; Cerda, A.; Dorea, E.L.; Pinto, G.A.; Gusukuma, M.C.; Bertolami, M.C.; Salazar, L.A.; Hirata, R.D.C. Impact of 3′UTR genetic variants in PCSK9 and LDLR genes on plasma lipid traits and response to atorvastatin in Brazilian subjects: A pilot study. Int. J. Clin. Exp. Med. 2015, 8, 5978–5988. [Google Scholar] [PubMed]
- Mahdy Ali, K.; Wonnerth, A.; Huber, K.; Wojta, J. Cardiovascular disease risk reduction by raising HDL cholesterol—Current therapies and future opportunities. Br. J. Pharmacol. 2012, 167, 1177–1194. [Google Scholar] [PubMed]
- Singh, I.M.; Shishehbor, M.H.; Ansell, B.J. High-density lipoprotein as a therapeutic target: A systematic review. JAMA 2007, 298, 786–798. [Google Scholar] [CrossRef] [PubMed]
- Nishizato, Y.; Ieiri, I.; Suzuki, H.; Kimura, M.; Kawabata, K.; Hirota, T.; Irie, S.; Kusuhara, H.; Urasaki, Y.; Akinori, U.; Urae, A.; et al. Polymorphisms of OATP-C (SLC21A6) and OAT3 (SLC22A8) genes: Consequences for pravastatin pharmacokinetics. Clin. Pharmacol. Ther. 2003, 73, 554–565. [Google Scholar] [CrossRef]
- Yang, G.P.; Yuan, H.; Tang, B.; Zhang, W.; Wang, L.S.; Huang, Z.J.; Ou-Yang, D.S.; Zhang, G.X.; Zhou, H.H. Lack of effect of genetic polymorphisms of SLCO1B1 on the lipid-lowering response to pitavastatin in Chinese patients. Acta Pharmacol. Sin. 2010, 31, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Pasanen, M.K.; Neuvonen, M.; Neuvonen, P.J.; Niemi, M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet. Genom. 2006, 16, 873–879. [Google Scholar] [CrossRef] [PubMed]
- Grundy, S.M.; Cleeman, J.I.; Merz, C.N.; Brewer, H.B., Jr.; Clark, L.T.; Hunninghake, D.B.; et al. Implications of recent clinical trials for the national cholesterol education program adult treatment panel III guidelines. Circulation 2004, 110, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo, P.C.; Munoz, M.A.; Lanas, M.C.; Lanas, Z.F.; Salazar, L.A. Endothelial nitric oxide synthase G894T gene polymorphism in Chilean subjects with coronary artery disease and controls. Clin. Chim. Acta 2006, 371, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Friedewald, W.T.; Levy, R.I.; Fredrickson, D.S. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 1972, 18, 499–502. [Google Scholar] [PubMed]
- Salazar, L.A.; Hirata, M.H.; Cavalli, S.A.; Machado, M.O.; Hirata, R.D. Optimized procedure for DNA isolation from fresh and cryopreserved clotted human blood useful in clinical molecular testing. Clin. Chem. 1998, 44, 1748–1750. [Google Scholar] [PubMed]
- Simon, J.A.; Lin, F.; Hulley, S.B.; Blanche, P.J.; Waters, D.; Shiboski, S.; Rotter, J.I.; Nickerson, D.A.; Yang, H.; Saad, M.; et al. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: The cholesterol and pharmacogenetics (CAP) Study. Am. J. Cardiol. 2006, 97, 843–850. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prado, Y.; Saavedra, N.; Zambrano, T.; Lagos, J.; Rosales, A.; Salazar, L.A. SLCO1B1 c.388A>G Polymorphism Is Associated with HDL-C Levels in Response to Atorvastatin in Chilean Individuals. Int. J. Mol. Sci. 2015, 16, 20609-20619. https://doi.org/10.3390/ijms160920609
Prado Y, Saavedra N, Zambrano T, Lagos J, Rosales A, Salazar LA. SLCO1B1 c.388A>G Polymorphism Is Associated with HDL-C Levels in Response to Atorvastatin in Chilean Individuals. International Journal of Molecular Sciences. 2015; 16(9):20609-20619. https://doi.org/10.3390/ijms160920609
Chicago/Turabian StylePrado, Yalena, Nicolás Saavedra, Tomás Zambrano, Jenny Lagos, Alexy Rosales, and Luis A. Salazar. 2015. "SLCO1B1 c.388A>G Polymorphism Is Associated with HDL-C Levels in Response to Atorvastatin in Chilean Individuals" International Journal of Molecular Sciences 16, no. 9: 20609-20619. https://doi.org/10.3390/ijms160920609