1. Introduction
The innate immune system acts as the front line of defense against invading microorganisms in most multicellular organisms by recognizing pathogen-associated molecular patterns (PAMPs) through pattern recognition receptors (PRRs) [
1]. Toll-like receptors (TLRs) form part of an ancient receptor family of PRRs and are the major cell-surface initiators of inflammatory responses to invading pathogens [
2]. To date, 13 TLRs have been identified in mammals and 10 have been found in avian species, including the chicken, goose, and duck [
3,
4,
5]. TLRs bind a wide variety of pathogen-associated substances through their ectodomains [
6]. The basic framework of TLRs is a horseshoe-shaped solenoid, or “m”-shaped architecture that contains an extensive β-sheet on its concave surface, and numerous ligand-binding insertions [
7]. These surfaces are responsible for ligand binding and ligand-induced TLR dimerization [
8].
TLRs comprise a family of type I transmembrane receptors [
9], which are composed of various leucine-rich repeats (LRRs), and are characterized by a signal peptide, an extracellular domain containing varying numbers of tandem LRRs, a
C-terminal capping region overlapping the last LRR (LRR-CT), a transmembrane helix, and a cytoplasmic region containing a Toll interleukin-1 receptor domain [
10,
11].
Induction of the innate immune pathways is critical for early anti-viral defense, but there is limited understanding of how pigeons recognize viral molecules and activate these pathways. In mammals, TLR7 recognizes single-stranded RNA of viral origin and synthetic anti-viral imidazoquinoline compounds [
12]. Upon activation, TLR7 recruits myeloid differentiation primary response protein 88 (MyD88) that, through several effector molecules, initiates the activation of two major signaling pathways resulting in the production of proinflammatory cytokines and/or type I interferons [
13]. Collectively, these observations indicate that TLR7 plays a role in restricting the entry of viral pathogens in mammals. However, in birds, such as pigeons,
TLR7 has not yet been cloned and its function has not been reported.
In this paper, we describe the cloning and characterization of the pigeon TLR7 sequence, analysis of its expression in various tissues, and evaluation of its function in response to the agonist R848 (resiquimod). Pigeon peripheral blood mononuclear cells (PBMCs) exposed to R848 stimulation showed significant induction of proinflammatory cytokine and interferon-γ (IFN-γ) production, but no significant change of the TLR7 mRNA level in vitro. After Newcastle disease virus (NDV) inoculation and R848 injection of pigeons, the expression levels of proinflammatory cytokines and IFN-γ in spleen were significantly up-regulated and TLR7 mRNA significantly increased in the R848-injected group, but significantly decreased in the LaSota-inoculated group at three day post-infection. These data expand our knowledge of the relationship between TLR7 and innate immunity in pigeons, which may be of relevance to other avian species, including goose and duck.
3. Discussion
Among the PRRs, TLRs are crucial for recognition of pathogen-derived products, and initiate signaling cascades leading to the activation of innate host defenses [
14]. In contrast to mammalian TLR7 orthologs, little is known about the sequence variation and biological function of avian TLR7. The demonstrated response of duck and goose TLR7 to imidazoquinoline suggests a similar role for avian TLR7 homologs in the immune response of birds [
15,
16].
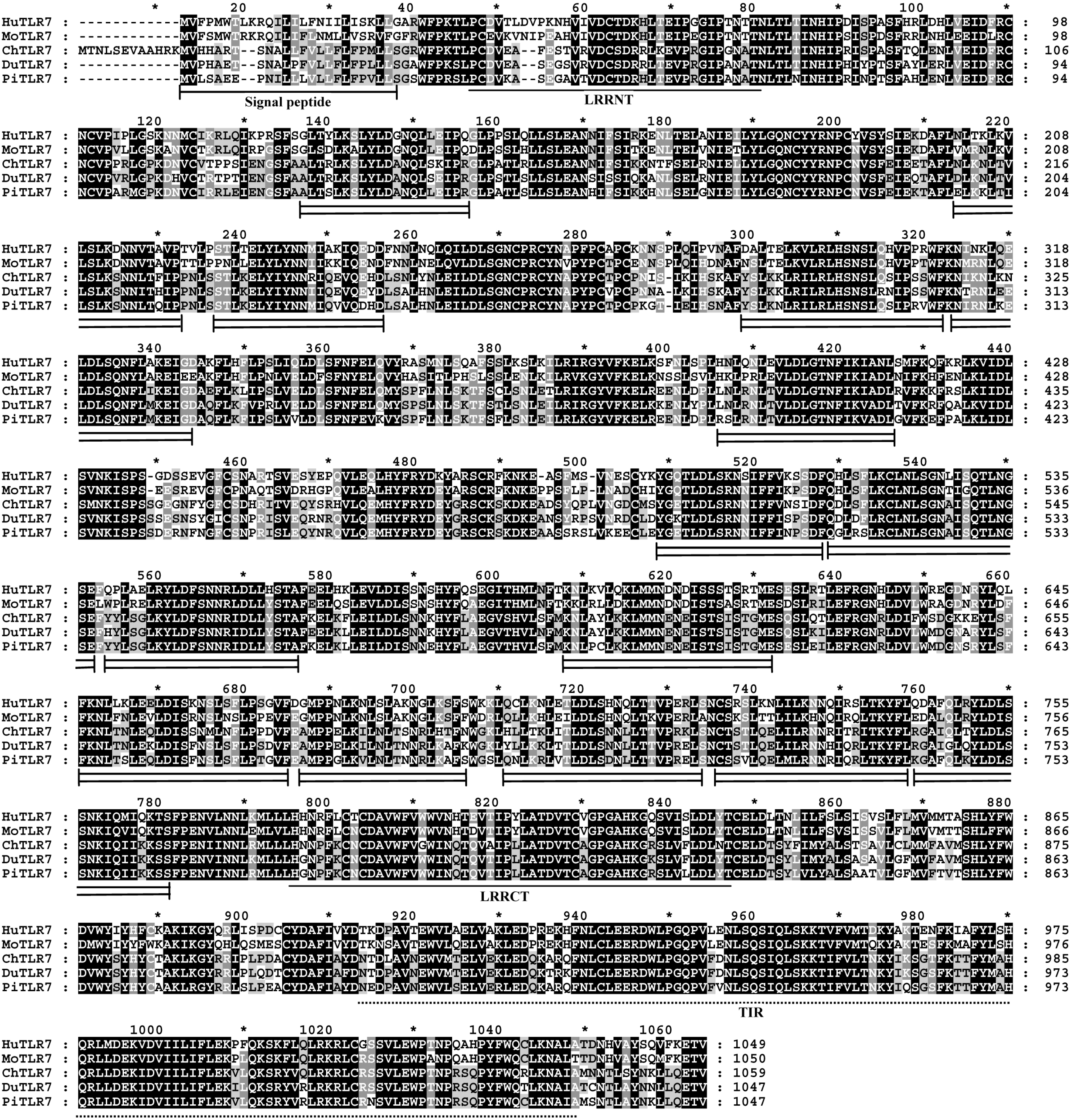
LRRs are found in a diverse set of proteins in which they are involved in ligand recognition and signal transduction [
17]. The structures of LRRs in TLRs are speculated to mediate recognition of specific PAMPs [
7]. Current evidence indicates that these irregular LRRs are involved in PAMP recognition, and a residue insertion after position 15 in LRR14 of human TLR5 contributes to flagellin binding [
18]. Interestingly, among the 15 LRR domains in pigeon TLR7 with the prevailing LRR motif XLXXLXLXXNXφXXφXXXXFXXLX [
10,
19], five LRRs differ from the canonical sequence by amino acid insertions, including LRR2, LRR11, LRR13, and LRR14 at position 15, and LRR10 at position 10. A
C-terminal proline-rich LRR, similar to some bacterial LRRs [
7], was found in LRR1, LRR2 and LRR11 at position 15 of pigeon TLR7 (
Figure 2). The most striking modifications to the TLR consensus are large insertions that occur at positions 10 or 15, and insertions or other irregularities on the convex surface might affect PAMP binding by introducing flexibility into the TLR horseshoe [
7]. Thus, the insertions in the LRRs of pigeon TLR7 may influence its functional role as a pathogen receptor and further studies are needed to investigate this.
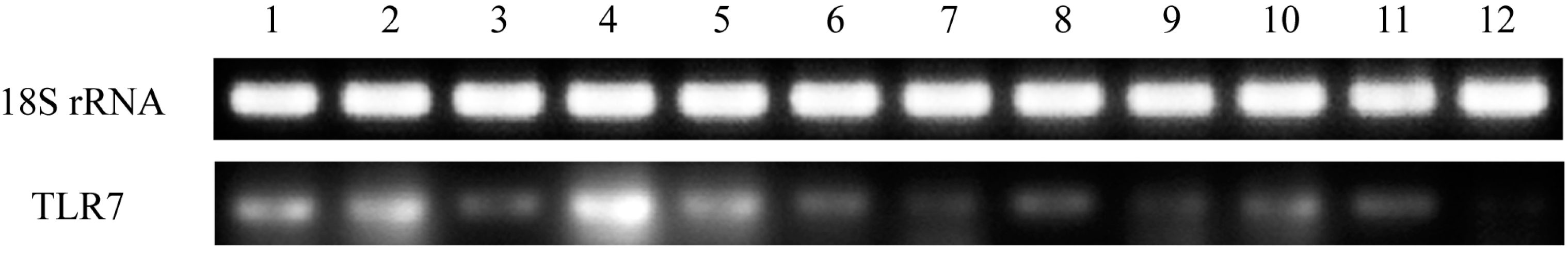
The gene expression patterns of
TLR7 in healthy pigeon tissues were assessed in this study. TLR7 was expressed in all tissues examined, although expression levels varied (
Figure 4). This is consistent with the finding that duck
TLR7 is expressed in a broad range of tissues and cell types [
15]. In this study, high expression levels of pigeon
TLR7 mRNA were observed in the PBMCs, spleen, liver, and lung, whereas the lowest expression was observed in the heart, small intestine, and brain. The expression profile of pigeon
TLR7 differs from that of goose
TLR7, which displays high expression levels in the cecum and lung, but low expression in the liver and kidney [
16]. These findings illustrate species-based differences in the tissue distribution of
TLR7 expression. The observation that pigeon
TLR7 is expressed at sites of viral contact in the lung and immune organs is consistent with a role in the host innate immune system as the first line of defense against pathogens in these tissues.
Imidazoquinoline is a TLR7 ligand [
20] that upon receptor binding with TLR7, induces translocation of NF-kB and production of TNF-α, IL-6, IL-12, and type I interferon [
12,
21].
In vitro experiments indicate that immunostimulatory RNA oligonucleotides (ORN) found in foot-and-mouth disease virus can activate nuclear factor-κB via porcine TLR7 [
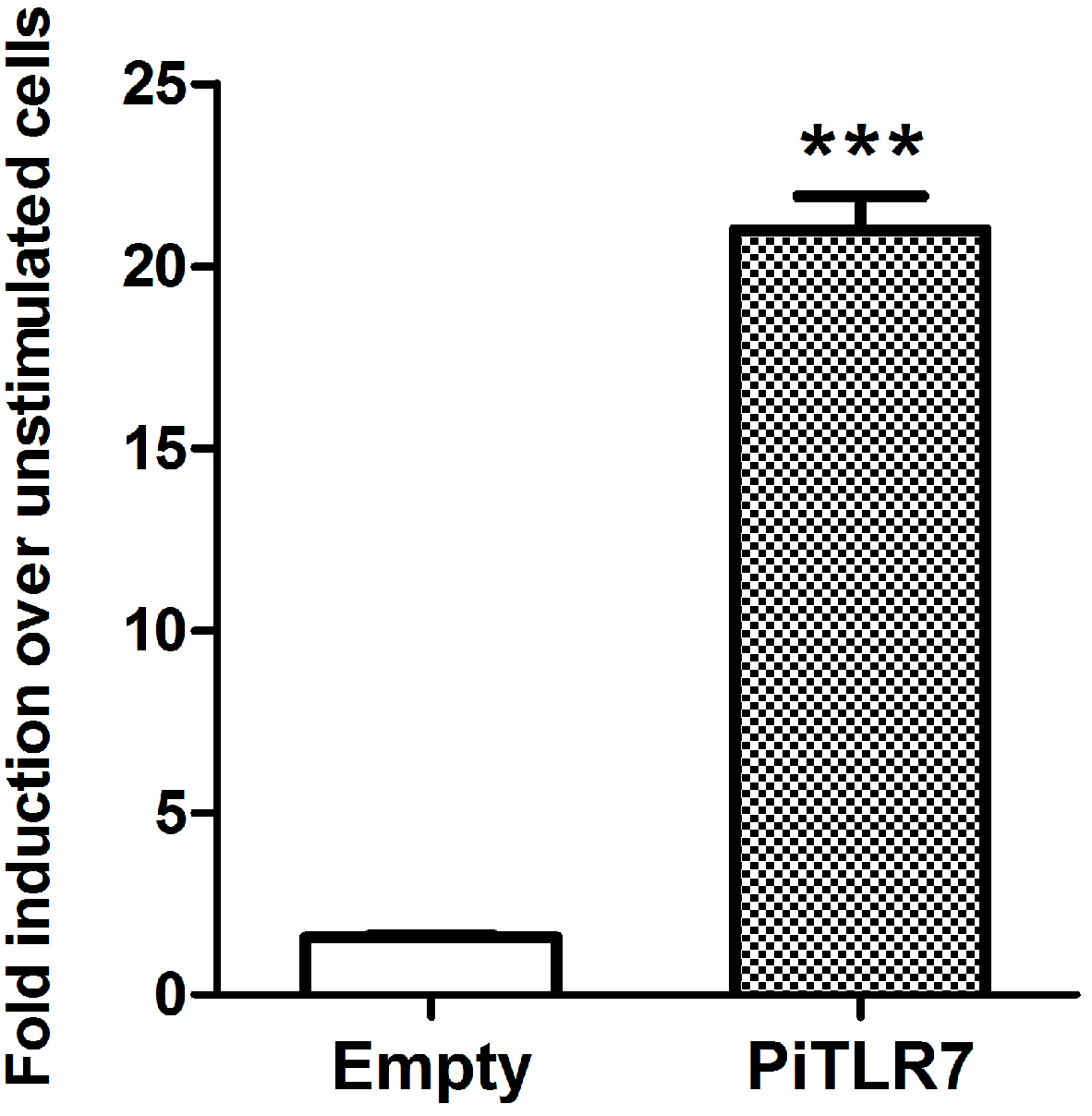
22]. In this study, following stimulation with R848, the induction level of NF-κB-induced luciferase activity in pigeon
TLR7-transfected HEK293T cells was significantly higher than that in cells transfected with an empty vector control (
Figure 6). Our results suggest that pigeon TLR7 is a pathogen receptor that plays a role in the recognition of the agonist R848.
Goose spleen mononuclear cells infected by New type gosling viral enteritis virus (NGVEV)
in vitro were unable to increase the expression of TLR7. Instead, levels of TLR7 in the spleen of NGVEV-infected geese increased dramatically [
16]. Similar to goose TLR7, the mRNA expression of pigeon
TLR7 was not statistically significantly different compared with controls following R848 stimulation of pigeon PBMCs (
Figure 7). However,
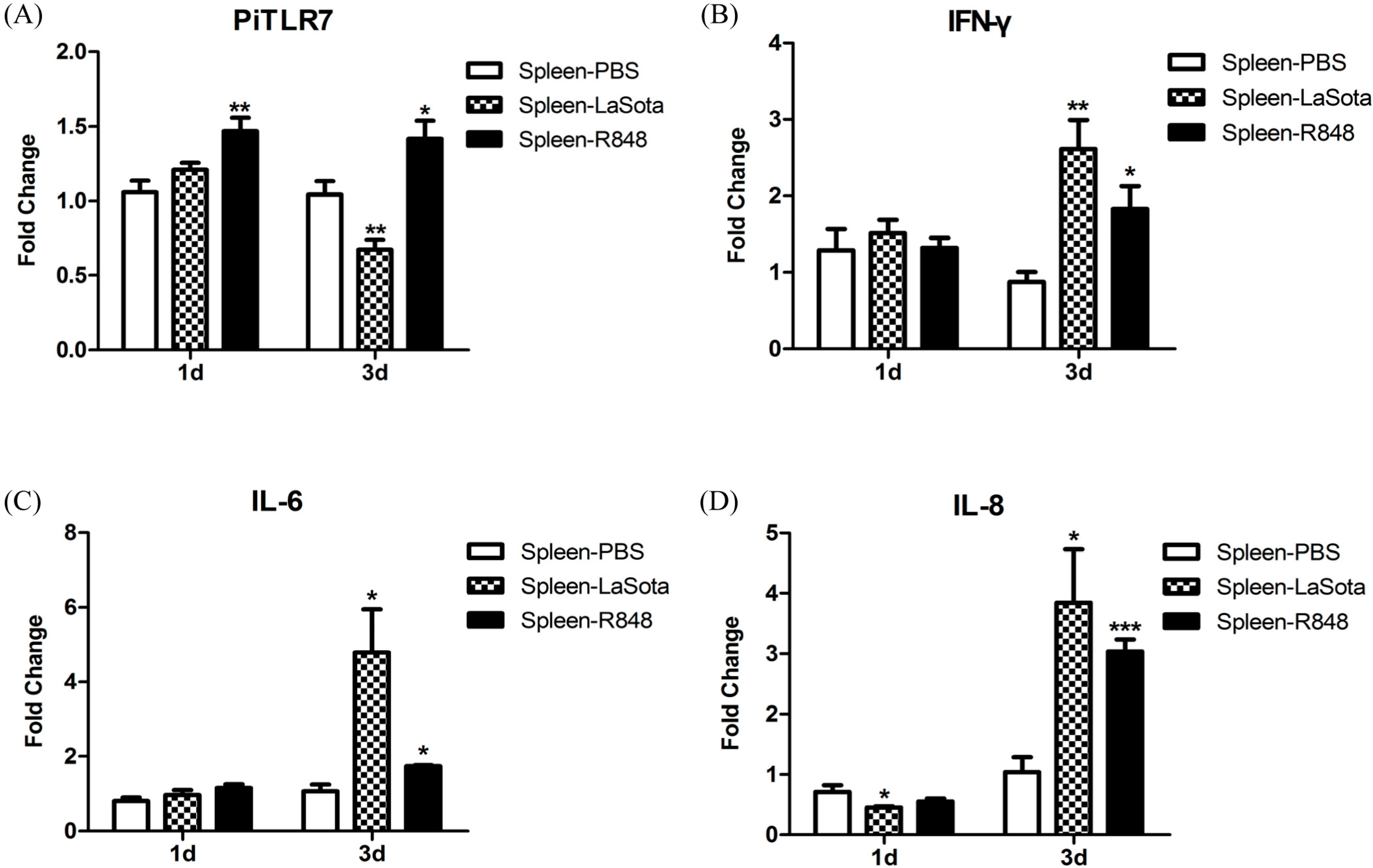
TLR7 gene expression in the spleen of pigeons injected with R848 was slightly but significantly increased over the controls (
Figure 8). Another study demonstrated that both duck and chicken
TLR7 are only transiently expressed in PBMCs at the early stages of low pathogenic avian influenza virus H11N9 infection, followed by a decline as the infection progresses [
23]. In variant strain infectious bursal disease virus-infected bursa, TLR7 expression was downregulated at 3 and 5 d.p.i. and upregulated at 7 d.p.i. [
24]. In this study, TLR7 expression in the LaSota-inoculated group appeared to be slightly elevated at 1 d.p.i., but this was not statistically significant, and significantly decreased 3 d.p.i. when compared with the control (
Figure 8).
TLR7 plays an essential role in inflammatory cytokine activation in granulocyte-macrophage colony-stimulating factor (GM-CSF)-primed murine neutrophils in response to influenza and the R848, as has been shown using
TLR7−/− mice. Murine
TLR7−/− neutrophils had poor responses to both R848 and influenza virus in comparison with wild-type cells [
25]. In this study, following R848 stimulation of pigeon PBMCs, the expression levels of the anti-viral cytokine IFN-γ, and of inflammatory cytokines, were robustly upregulated at 12 and 24 h.p.i. Interestingly, the anti-inflammatory cytokine IL-10 was also significantly increased at both time points (
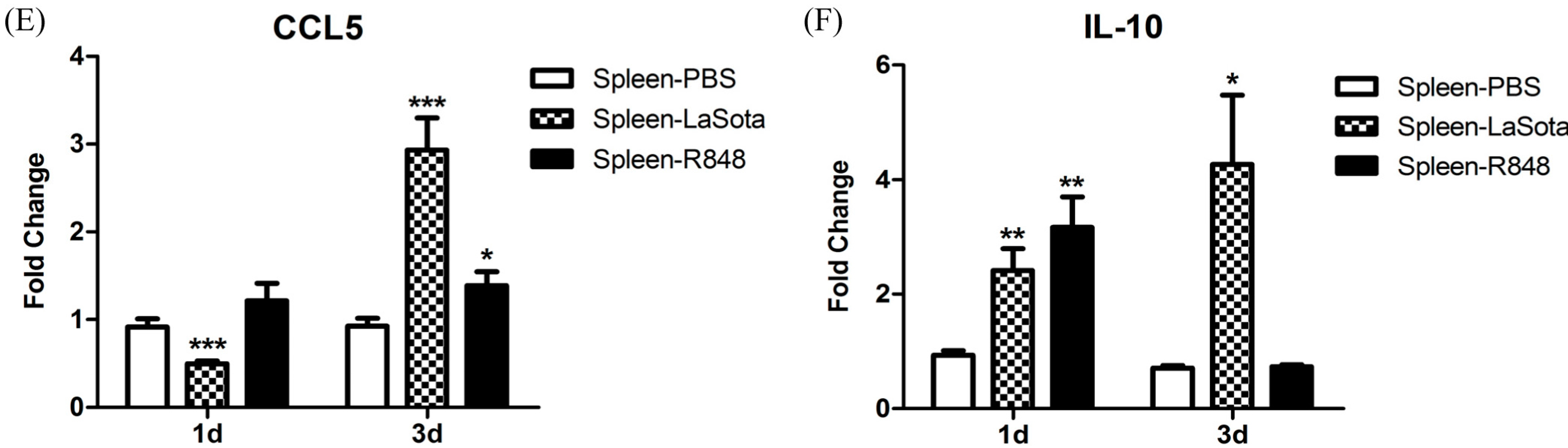
Figure 7) and further studies are needed to ascertain the reason for this observation. In response to LaSota inoculation or R848 injection of pigeons, the mRNA levels of
IFN-γ,
IL-6,
IL-8,
CCL5, and
IL-10 in the spleen were markedly elevated (
Figure 8). An
in vivo study by Rasoli
et al. reported that significant upregulation in the mRNA expression level of IL10 correlated with downregulation of the mRNA levels of proinflammatory cytokines and chemokines in AF2240 and IBS002 strains of NDV-infected spleen at 4 d.p.i. [
26]. Notably, we observed that the level of the anti-inflammatory cytokine IL-10 was dramatically reduced (to the original level in the R848-injected group) to 3 d.p.i., which was concomitant with the upregulation of inflammatory cytokines (
Figure 8). R848 stimulation or LaSota inoculation resulted in the rapid up-regulation of proinflammatory cytokines and anti-viral molecules, suggesting that pigeon TLR7 plays an important role in the innate immune response.
4. Experimental Section
4.1. Molecular Cloning of Pigeon TLR7
King pigeons (
Columba livia) purchased from Jiangyin Wei Tekai Pigeon Co. (Wuxi, China) were housed in isolators and fed with a pathogen-free diet and water. The procedures described in this study were approved by the Ethics Committee on Animal Experiments of Yangzhou University, Yangzhou, China (Yangzhou University, [2012] no. 62, 12 December 2012). Genomic DNA was extracted from whole blood of the pigeons using a Universal Genomic DNA Extraction Kit Ver. 3.0 (Takara Biotechnology Co., Dalian, China) according to the manufacturer’s instructions. To clone the pigeon
TLR7 gene, primer pairs covering the entire open reading frame (ORF) were designed based on the predicted pigeon nucleotide (GenBank ID: XM_005512700) and the well-conserved upstream and downstream sequences of
TLR7 from chicken (
Gallus gallus; GenBank ID: NM_001011688) and duck (
Anas platyrhynchos; GenBank ID: DQ888645). PCR was performed using the designed primers (
Table 1) and pigeon genomic DNA as a template. PCR was carried out using PrimeSTAR HS DNA Polymerase (Takara Biotechnology Co.) in a 50-μL reaction volume consisting of 1× PrimeSTAR Buffer (Mg
2+ plus), 200 µM of each deoxy-ribonucleoside triphosphate (dNTP), 0.2 µM of forward and reverse primers, 1.25 U of PrimeSTAR Hot Start (HS) DNA Polymerase, and ~200 ng of genomic DNA template. PCR amplifications were performed as follows: 1 cycle of 98 °C for 5 min, then 30 cycles of 98 °C for 10 s, 60 °C for 15 s, and 72 °C for 3 min, followed by 1 cycle of 72 °C for 10 min. The amplified PCR product was purified, cloned into pCR2.1-T using a TA Cloning Kit (Invitrogen, Carlsbad, CA, USA), and sequenced by Genscript (Nanjing, China).
Table 1.
PCR primers used in this study. The underlined parts in PiTLR7 F/R represent the introduced SalI and KpnI restriction enzyme sites.
Table 1.
PCR primers used in this study. The underlined parts in PiTLR7 F/R represent the introduced SalI and KpnI restriction enzyme sites.
| Primer Name | Primer Sequence (5'→3') | Size (bp) | Application |
|---|
| PiTLR7 F | ACGCGTCGACCATGGTACTTAGTGCAGAAGAGCCAAATAC | 3144 | Amplification of Pigeon TLR7 ORF |
| PiTLR7 R | CCCCGGTACCCTAAACAGTTTCTTGGAGAAGCTTGTTG |
| PiTLR7-RT-F | CAGACTCAAGTGACTATTCCTCTTCTG | 216 | RT-PCR |
| PiTLR7-RT-R | GTAACTATACCACACATCCCAGAAATAGAG |
| 18S rRNA F | TTGGTGGAGCGATTTGTC | 129 | RT-PCR |
| 18S rRNA R | ATCTCGGGTGGCTGAACG |
| Piβ-actin F | ATGAAGCCCAGAGCAAAAGAG | 223 | Quantitative real-time PCR |
| Piβ-actin R | GGGGTGTTGAAGGTCTCAAAC |
| PiTLR7 F | ACCAGCGGCTTCTAGATGAA | 158 | Quantitative real-time PCR |
| PiTLR7 R | CTGCCAAAAGTAGGGCTGAG |
| PiIFN-γ F | CAGACGTAGCTGATGGTGGAC | 233 | Quantitative real-time PCR |
| PiIFN-γ R | AAGCTTTGCCAGATCCTTGAG |
| PiIL-6 F | AGCGTCGATTTGCTGTGCT | 107 | Quantitative real-time PCR |
| PiIL-6 R | GATTCCTGGGTAGCTGGGTCT |
| PiIL-8 F | CTGTCCTGGCTCTTTTCCTG | 199 | Quantitative real-time PCR |
| PiIL-8 R | CTGCCGTCCTTCAGAGTAGC |
| PiCCL5 F | GTGAAGGACTATTTCTACACCAGCA | 95 | Quantitative real-time PCR |
| PiCCL5 R | GCGTCAGGGTTTGCACAGA |
| PiIL-10 F | TGATGAACTTAGCATCCAGCTACTC | 93 | Quantitative real-time PCR |
| PiIL-10 R | AACTGCATCATCTCCGACACA |
4.2. Sequence Analyses
The nucleotide and deduced amino acid sequences of pigeon
TLR7 were analyzed using DNAstar software and the Expert Protein Analysis System (Expasy,
http://www.mrc-lmb.cam.ac.uk/genomes/madanm/pres/swiss1.htm). SMART was used to predict the protein domain structure of pigeon
TLR7.
TLR7 sequences from different species were compared using the NCBI BLAST search program (
http://blast.ncbi.nlm.nih.gov/Blast.cgi). A multiple sequence alignment was performed using ClustalW (
http://www.ebi.ac.uk/clustalw/) and edited with the Genedoc program. Phylogenetic analysis was conducted on amino acid sequences using MEGA 5.1 software (
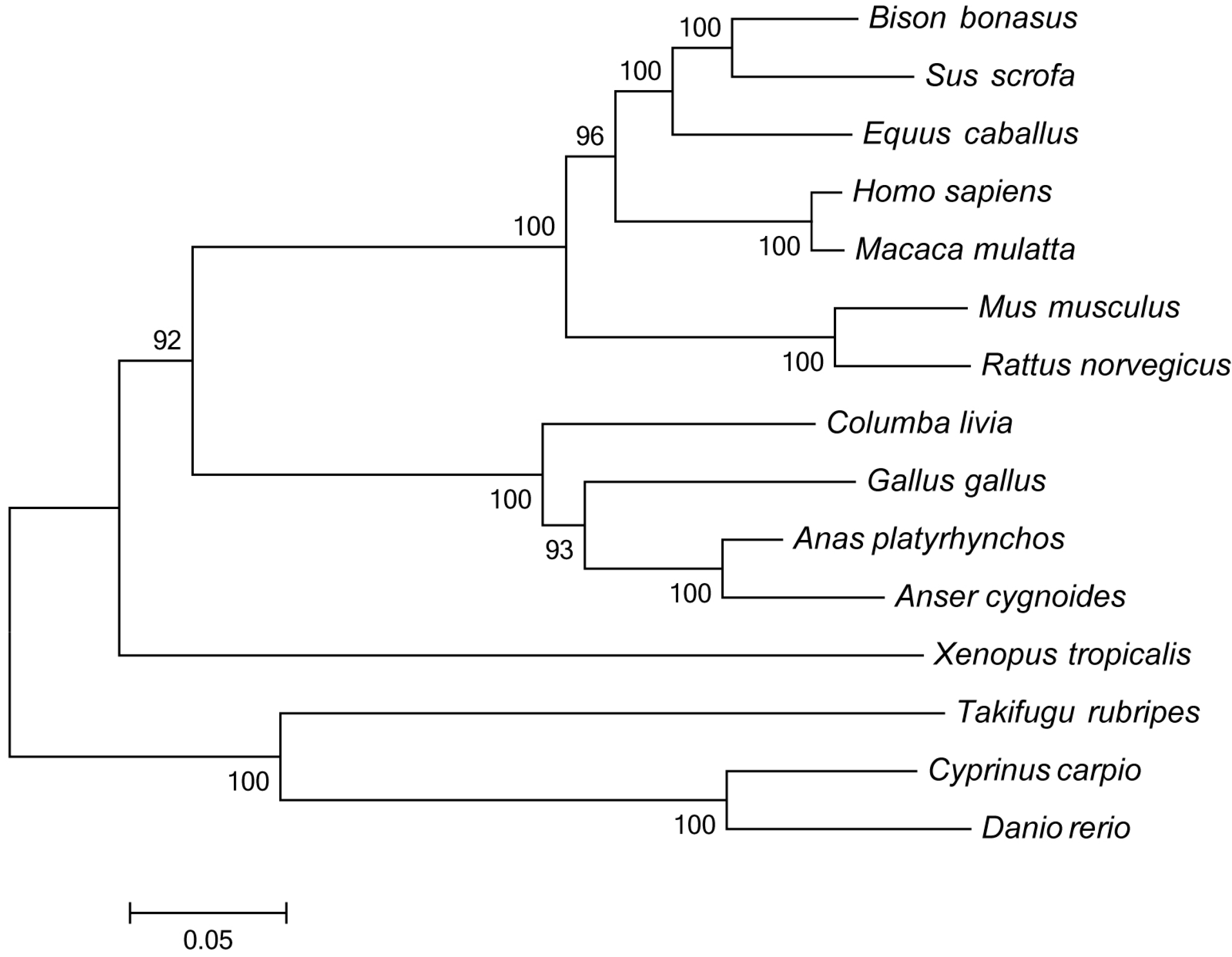
http://www.megasoftware.net), and a phylogenetic tree was constructed with the neighbor-joining method using a Poisson correction model with 1000 bootstrap replicates.
4.3. Semi-Quantitative Analysis of Pigeon TLR7 Expression in Tissues
Total RNA was extracted from peripheral blood mononuclear cells (PBMCs), spleen, neck lymph nodes, liver, lung, kidney, heart, bone marrow, small intestine, large intestine, cecum, and brain of healthy King pigeons using TRIzol reagent (Invitrogen), according to the manufacturer’s instructions. Total RNA samples were treated with RNase-Free DNase I (Takara Biotechnology Co.) to remove contaminating DNA, and first-strand cDNA was synthesized using M-MLV reverse transcriptase (Invitrogen) following the manufacturer’s protocol. RT-PCR was performed with cDNA templates from each tissue and a specific primer set for
TLR7 (
Table 1). The endogenous control was 18S rRNA from pigeon (GenBank ID: AF173630). PCR products were run on 1% (
w/
v) agarose gels.
4.4. DNA Constructs
An expression construct, pCMV-TLR7, was made by cloning the full-length pigeon TLR7 coding sequence into the SalI and KpnI sites of the pCMV-HA-tag expression vector (Invitrogen). Plasmid pCR2.1-TLR7, containing the full-length TLR7 open reading frame (ORF), was digested with SalI and KpnI (Takara Biotechnology Co.), and subcloned into the same restriction enzymes sites of pCMV-HA. The resulting construct was designated pCMV-PiTLR7.
4.5. Cell Culture, Transfection and Stimulation
HEK293T cells were grown in 24-well tissue culture plates in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) until 70% confluence was reached. Cells were washed with PBS before transfection, and the medium was then replaced with Opti-MEM (Invitrogen) medium. Transient transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Equal amounts of DNA constructs, including pCMV-TLR7 or pCMV-empty, and 400 ng of reporter pGL4.32 (luc2P/NF-κB-RE/Hygro) plasmid DNA (Promega, Madison, WI, USA) were transfected into HEK293T cells. Twenty-four hours after transfection, endotoxin-free R848 (Enzo Life Sciences, Farmingdale, NY, USA) was added to a final concentration of 2.5 μg/mL. After stimulation for 5 h, cells were harvested for subsequent western blotting or Luciferase assay.
4.6. Western Blotting Analysis
Harvested cells were subjected to SDS-PAGE and the proteins were transferred to a nitrocellulose membrane. The membrane was blocked with blocking buffer (5% non-fat dry milk and 0.05% Tween-20 in PBS) at 4 °C overnight. The next day, the membrane was incubated with an anti-TLR7 monoclonal antibody raised against a partial recombinant human TLR7 (NP_057646, 27~126 amino acids, sharing 77% similarity with pigeon TLR7) (1:800 diluted in PBS-0.05% Tween 20, Abnova Corporation, Walnut, CA, USA) at 37 °C for 2 h. After washing three times with PBS-0.05% Tween 20 (PBST), the membrane was incubated with a horseradish peroxidase-conjugated secondary antibody (1:5000 diluted in PBST, Sigma, St. Louis, MO, USA) at 37 °C for 1 h. The protein bands were visualized using an enhanced chemiluminescence reagent (Thermo, Rockford, IL, USA).
4.7. Luciferase Assay
To determine the functional response of pigeon TLR7 to R848 stimulation, NF-κB-induced luciferase activity was measured using the Bright-Glo Luciferase Assay system (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
4.8. Pigeon PBMCs Preparation and Stimulation
PBMCs were isolated from whole blood as previously described [
27]. Briefly, the blood sample was collected with 10% EDTA at a 10:1 (
v:
v) ratio from healthy King pigeons and centrifuged for 10 min at 200×
g. The erythrocyte pellet was resuspended in PBS, then layered onto 1077 Histopaque (Sigma) and centrifuged at 400×
g for 30 min. Mononuclear cells were collected from the gradient interface, washed with PBS, and centrifuged for 10 min at 200×
g. A final wash was performed with antibiotic-free RPMI 1640 (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Invitrogen). The pellets were then resuspended in antibiotic-free RPMI 1640 with FBS. Cell viability and number were determined by trypan blue exclusion. Pigeon PBMCs were cultured in RPMI 1640 medium containing 2% FBS, 1% penicillin/streptomycin, and 2 mM
l-glutamine. After plating, cells were incubated overnight at 41 °C and then washed with PBS to remove non-adherent cells. Cells were stimulated with the TLR7 agonist R848 at a concentration of 2.5 μg/mL for either 12 or 24 h, then harvested for subsequent mRNA detection of immune-related genes.
4.9. Virus Inoculation and R848 Injection of Pigeons
King pigeons were housed and handled following approval by the Institutional Animal Experimental Committee (Yangzhou University, [2012] no. 62, 12 December 2012). Pigeons were randomly divided into three groups (six pigeons per group) and injected via an intramuscular route with either 10 μg R848 (Enzo Life Sciences, Farmingdale, NY, USA), 106 EID50 of LaSota (Wuhan Chopper Biology Co., Wuhan, China), or PBS in 200 μL. At 1 and 3 d.p.i., pigeons were sacrificed by pentobarbital overdose, and the spleens were removed and stored at −70 °C for subsequent mRNA analysis of immune-related genes.
4.10. RNA Isolation, RT-PCR and Quantitative Real-Time PCR
Harvested cells or spleens were homogenized in TRIzol reagent (Invitrogen), and total RNA was prepared as described by the manufacturer. RNA concentrations were determined by spectrophotometer readings at 260 nm. Quantitative real-time PCR (qRT-PCR) was performed to measure mRNA expression levels of
TLR7,
IFN-γ,
IL-6,
IL-8,
CCL5, and
IL-10 using SYBR Premix Ex Taq II (Perfect Real Time; Takara Biotechnology Co.) using an ABI 7500 real-time detection system (Applied Biosystems, Carlsbad, CA, USA) with designed primers (
Table 1). Amplification was performed in a total volume of 20 μL containing 10 μL of 2× SYBR Premix Ex Taq II, 2 μL of the diluted cDNA, and 0.8 μL of each primer. The real-time PCR program started with denaturing at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. Dissociation analysis of amplification products was performed at the end of each PCR to confirm that only one PCR product was amplified and detected. Data were analyzed using ABI 7500 SDS software (Applied Biosystems, Foster, CA, USA), with the baseline being set automatically by the software. The threshold method was used for quantification of the mRNA level [
28] and Δ
Ct values were calculated on the basis of the internal standard (ACTB, actin-beta) signal. Results are expressed as 2
−ΔΔCt (n-fold change compared to the control group).
4.11. Statistical Analysis
The significance of the differences in the experimental data between TLR7-positive and empty vector-transfected groups, or between R848-stimulated/LaSota-inoculated and control groups, were determined using the Student’s t-test with Instat version 5.0 (GraphPad Software, San Diego, CA, USA). Statistical significance was determined at p < 0.05 (*), p < 0.01 (**) or p < 0.001 (***).