Residue Asn277 Affects the Stability and Substrate Specificity of the SMG1 Lipase from Malassezia globosa
Abstract
:1. Introduction
2. Results and Discussion
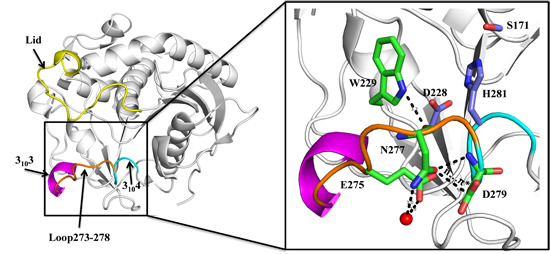
2.1. N277 Stabilizing Loop 273–278 and 3104 Helix
2.2. Biochemical Properties of SMG1 Mutants
2.2.1. Effect of Temperature and pH
| Mutant | Topt (°C) | pHopt | Substrateopt | Thermostability (%) a | Specific Activity (U/g) b | Tm (°C) c |
|---|---|---|---|---|---|---|
| SMG1 WT | 25 | 6 | pNP caprylate | 67.7 | 15,425.4 | 54.6 |
| N277D | 20 | 6 | pNP caprylate | 78.7 | 16,081.8 | 56.6 |
| N277V | 15 | 6 | pNP caprylate | 38.9 | 5292.2 | 41.1 |
| N277L | 20 | 6 | pNP caprylate | 33.5 | 4320.5 | 42.6 |
| N277F | 20 | 6 | pNP caprylate | 0 | 535.2 | 38.3 |

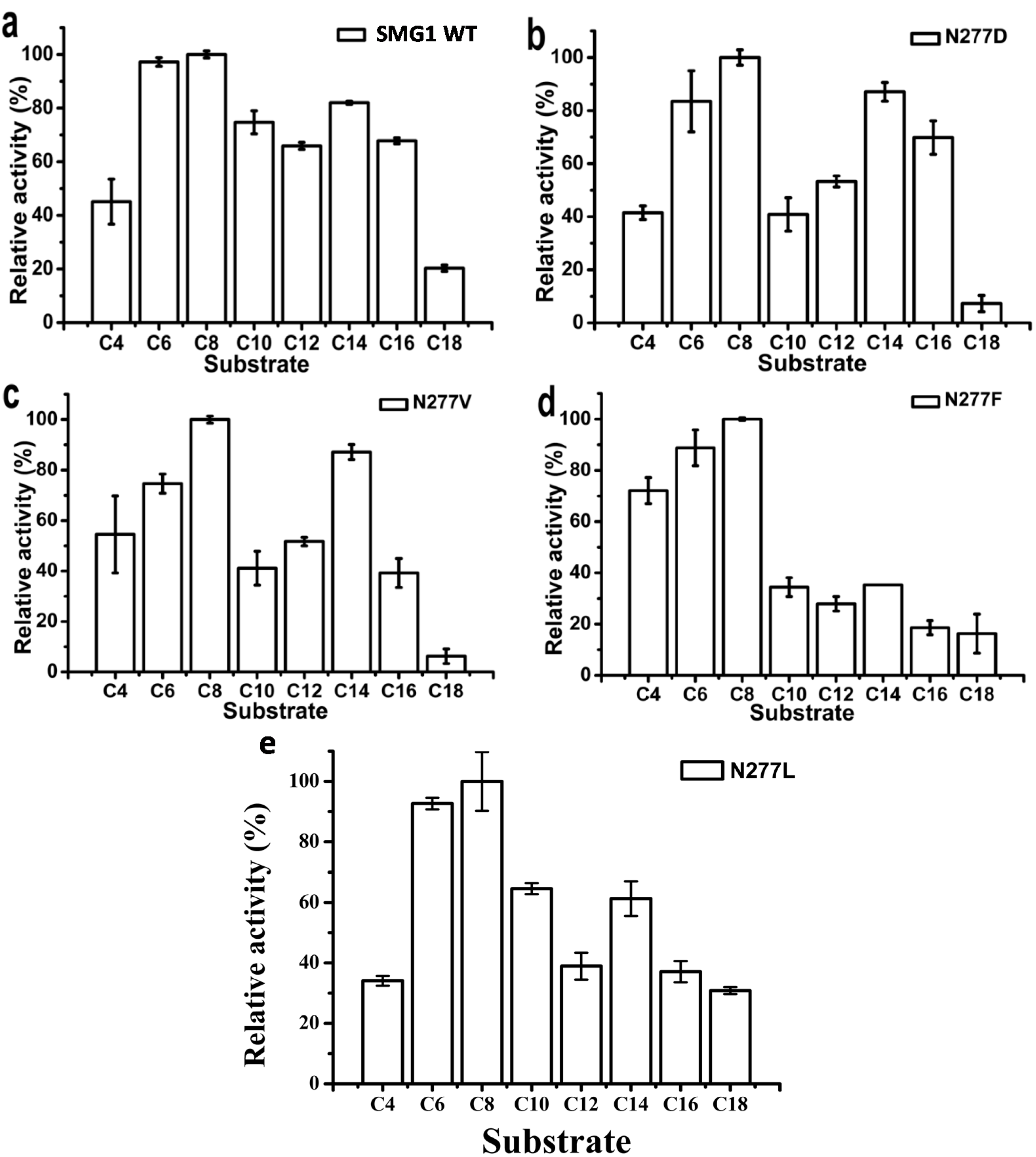
2.2.2. Substrate Specificity of Artificial pNP Esters
2.2.3. Substrate Specificity of Acylglycerols

| Mutants | Δ1,3-DAG (%) | Δ1,2-DAG (%) | Δ1,3-DAG/Δ1,2-DAG |
|---|---|---|---|
| SMG1 WT | 27.25 | 14.37 | 1.90 |
| N277D | 27.09 | 14.18 | 1.91 |
| N277V | 26.68 | 13.35 | 2.00 |
| N277L | 26.73 | 12.83 | 2.08 |
| N277F | 10.1 | 7.78 | 1.30 |
2.2.4. Fatty Acids Selectivity of Mutants
| Fatty Acids | SMG1 WT | N277D | N277L | N277V | N277F |
|---|---|---|---|---|---|
| C6:0 | 0.28 | 0.28 | 0.91 | 0.22 | 0.40 |
| C8:0 | 0.64 | 0.64 | 0.59 | 0.29 | 0.94 |
| C10:0 | 0.91 | 0.90 | 0.50 | 0.71 | 1.00 |
| C12:0 | 0.82 | 0.83 | 0.46 | 1.00 | 0.85 |
| C14:0 | 0.95 | 0.96 | 0.55 | 0.90 | 0.64 |
| C16:0 | 1.00 | 1.00 | 1.00 | 0.98 | 0.72 |
| C18:0 | 0.97 | 0.97 | 0.81 | 0.97 | 0.52 |
2.3. Molecular Basis of SMG1 Mutants

3. Experimental Section
3.1. Bacterial Strains, Chemicals, Plasmids and Medium
3.2. Expression and Purification of SMG1 and Its Mutants
3.3. Biochemical Properties of Recombinant Lipases
3.4. Circular Dichroism Spectral Analysis
3.5. Hydrolysis of Acylglycerols
3.6. Fatty Acid Selectivity Determination
3.6.1. Esterification
3.6.2. Gas Chromatography Analysis
3.6.3. Determination of the Competitive Factor
3.7. 3-D Structure Models of SMG1 Mutants
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Houde, A.; Kademi, A.; Leblanc, D. Lipases and their industrial applications—An overview. Appl. Biochem. Biotechnol. 2004, 118, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, K.E.; Eggert, T. Lipases for biotechnology. Curr. Opin. Biotechnol. 2002, 13, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Verger, R. “Interfacial activation” of lipases: Facts and artifacts. Trends Biotechnol. 1997, 15, 32–38. [Google Scholar] [CrossRef]
- Rehm, S.; Trodler, P.; Pleiss, J. Solvent-induced lid opening in lipases: A molecular dynamics study. Protein Sci. 2010, 19, 2122–2130. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Lorente, G.; Cabrera, Z.; Godoy, C.; Fernandez-Lafuente, R.; Palomo, J.M.; Guisan, J.M. Interfacially activated lipases against hydrophobic supports: Effect of the support nature on the biocatalytic properties. Process Biochem. 2008, 43, 1061–1067. [Google Scholar] [CrossRef]
- Manoel, E.A.; Dos Santos, J.C.; Freire, D.M.; Rueda, N.; Fernandez-Lafuente, R. Immobilization of lipases on hydrophobic supports involves the open form of the enzyme. Enzym. Microb. Technol. 2015, 71, 53–57. [Google Scholar] [CrossRef]
- Fernandez-Lorente, G.; Palomo, J.M.; Cabrera, Z.; Fernandez-Lafuente, R.; Guisan, J.M. Improved catalytic properties of immobilized lipases by the presence of very low concentrations of detergents in the reaction medium. Biotechnol. Bioeng. 2007, 97, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Akoh, C.C.; Lee, G.C.; Shaw, J.F. Protein engineering and applications of Candida rugosa lipase isoforms. Lipids 2004, 39, 513–526. [Google Scholar] [CrossRef] [PubMed]
- Bornscheuer, U.T.; Bessler, C.; Srinivas, R.; Krishna, S.H. Optimizing lipases and related enzymes for efficient application. Trends Biotechnol 2002, 20, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.T.; Qi, F.; Yuan, C.; Zhao, X.; Ramkrishna, D.; Liu, D.; Varma, A. Lipase-catalyzed process for biodiesel production: Protein engineering and lipase production. Biotechnol. Bioeng. 2014, 111, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Santarossa, G.; Lafranconi, P.G.; Alquati, C.; DeGioia, L.; Alberghina, L.; Fantucci, P.; Lotti, M. Mutations in the “lid” region affect chain length specificity and thermostability of a Pseudomonas fragi lipase. FEBS Lett. 2005, 579, 2383–2386. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; An, J.; Yang, G.; Wu, G.; Zhang, Y.; Cui, L.; Feng, Y. Enhanced enzyme kinetic stability by increasing rigidity within the active site. J. Biol. Chem. 2014, 289, 7994–8006. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T.; Bocola, M.; Carballeira, J.D.; Zha, D.X.; Vogel, A. Expanding the range of substrate acceptance of enzymes: Combinatorial active-site saturation test. Angew. Chem. Int. Ed. 2005, 44, 4192–4196. [Google Scholar] [CrossRef]
- Garcia-Galan, C.; Berenguer-Murcia, A.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzym. Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Wang, W.F.; Li, T.; Qin, X.L.; Ning, Z.X.; Yang, B.; Wang, Y.H. Production of lipase SMG1 and its application in synthesizing diacylglyecrol. J. Mol. Catal. B-Enzym. 2012, 77, 87–91. [Google Scholar] [CrossRef]
- Zheng, P.Y.; Xu, Y.; Wang, W.F.; Qin, X.L.; Ning, Z.X.; Wang, Y.H.; Yang, B. Production of diacylglycerol-mixture of regioisomers with high purity by two-step enzymatic reactions combined with molecular distillation. J. Am. Oil Chem. Soc. 2014, 91, 251–259. [Google Scholar] [CrossRef]
- Xu, D.; Sun, L.J.; Chen, H.Y.; Lan, D.M.; Wang, Y.H.; Yang, B. Enzymatic synthesis of diacylglycerols enriched with conjugated linoleic acid by a novel lipase from Malassezia globosa. J. Am. Oil Chem. Soc. 2012, 89, 1259–1266. [Google Scholar]
- Nagao, T.; Watanabe, H.; Goto, N.; Onizawa, K.; Taguchi, H.; Matsuo, N.; Yasukawa, T.; Tsushima, R.; Shimasaki, H.; Itakura, H. Dietary diacylglycerol suppresses accumulation of body fat compared to triacylglycerol in men in a double-blind controlled trial. J. Nutr. 2000, 130, 792–797. [Google Scholar] [PubMed]
- Flickinger, B.D.; Matsuo, N. Nutritional characteristics of DAG oil. Lipids 2003, 38, 129–132. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.T.; Liu, L.; Hou, S.L.; Xu, J.X.; Yang, B.; Wang, Y.H.; Liu, J.S. Crystal structure of a mono- and diacylglycerol lipase from Malassezia globosa reveals a novel lid conformation and insights into the substrate specificity. J. Struct. Biol. 2012, 178, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Gao, C.L.; Lan, D.M.; Yang, B.; Wang, Y.H. Molecular basis for substrate selectivity of a mono- and diacylglycerol lipase from Malassezia globosa. Biochem. Biophys. Res. Commun. 2012, 424, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Lan, D.; Liu, L.; Zhang, H.; Yang, B.; Wang, Y. Site-directed mutagenesis studies of the aromatic residues at the active site of a lipase from Malassezia globosa. Biochimie 2014, 102, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, G.J.; Newberry, R.W.; van Veller, B.; Raines, R.T.; Woolfson, D.N. Interplay of hydrogen bonds and n→π* interactions in proteins. J. Am. Chem. Soc. 2013, 135, 18682–18688. [Google Scholar] [CrossRef] [PubMed]
- McGaughey, G.B.; Gagne, M.; Rappe, A.K. π-Stacking interactions. Alive and well in proteins. J. Biol. Chem. 1998, 273, 15458–15463. [Google Scholar]
- Li, A.J.; Nussinov, R. A set of van der Waals and coulombic radii of protein atoms for molecular and solvent-accessible surface calculation, packing evaluation, and docking. Proteins 1998, 32, 111–127. [Google Scholar] [CrossRef] [PubMed]
- Hendsch, Z.S.; Tidor, B. Do salt bridges stabilize proteins?A continuum electrostatic analysis. Protein Sci. 1994, 3, 211–226. [Google Scholar]
- Baxa, M.C.; Haddadian, E.J.; Jumper, J.M.; Freed, K.F.; Sosnick, T.R. Loss of conformational entropy in protein folding calculated using realistic ensembles and its implications for NMR-based calculations. Proc. Natl. Acad. Sci. USA 2014, 111, 15396–15401. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.F.; Li, T.; Ning, Z.X.; Wang, Y.H.; Yang, B.; Ma, Y.J.; Yang, X.Q. A process for the synthesis of PUFA-enriched triglycerides from high-acid crude fish oil. J. Food Eng. 2012, 109, 366–371. [Google Scholar] [CrossRef]
- Qin, X.L.; Lan, D.M.; Zhong, J.F.; Liu, L.; Wang, Y.H.; Yang, B. Fatty acid specificity of T1 lipase and its potential in acylglycerol synthesis. J. Sci. Food Agric. 2014, 94, 1614–1621. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.K.; Yang, B.; Wang, Y.H.; Wang, W.F.; Ning, Z.X.; Li, L. Enzymatic production of monoacylglycerols with camellia oil by the glycerolysis reaction. J. Am. Oil Chem. Soc. 2010, 87, 531–537. [Google Scholar] [CrossRef]
- Rangheard, M.S.; Langrand, G.; Triantaphylides, C.; Baratti, J. Multi-competitive enzymatic reactions in organic media: A simple test for the determination of lipase fatty acid specificity. Biochim. Biophys. Acta 1989, 1004, 20–28. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific LLC: Palo Alto, CA, USA. Available online: http://www.pymol.org (accessed on 11 December 2014).
- Wang, W.F.; Xu, Y.; Qin, X.L.; Lan, D.M.; Yang, B.; Wang, Y.H. Immobilization of lipase SMG1 and its application in synthesis of partial glycerides. Eur. J. Lipid Sci. Technol. 2014, 116, 1063–1069. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lan, D.; Wang, Q.; Xu, J.; Zhou, P.; Yang, B.; Wang, Y. Residue Asn277 Affects the Stability and Substrate Specificity of the SMG1 Lipase from Malassezia globosa. Int. J. Mol. Sci. 2015, 16, 7273-7288. https://doi.org/10.3390/ijms16047273
Lan D, Wang Q, Xu J, Zhou P, Yang B, Wang Y. Residue Asn277 Affects the Stability and Substrate Specificity of the SMG1 Lipase from Malassezia globosa. International Journal of Molecular Sciences. 2015; 16(4):7273-7288. https://doi.org/10.3390/ijms16047273
Chicago/Turabian StyleLan, Dongming, Qian Wang, Jinxin Xu, Pengfei Zhou, Bo Yang, and Yonghua Wang. 2015. "Residue Asn277 Affects the Stability and Substrate Specificity of the SMG1 Lipase from Malassezia globosa" International Journal of Molecular Sciences 16, no. 4: 7273-7288. https://doi.org/10.3390/ijms16047273
APA StyleLan, D., Wang, Q., Xu, J., Zhou, P., Yang, B., & Wang, Y. (2015). Residue Asn277 Affects the Stability and Substrate Specificity of the SMG1 Lipase from Malassezia globosa. International Journal of Molecular Sciences, 16(4), 7273-7288. https://doi.org/10.3390/ijms16047273