Identification and Functional Analysis of MicroRNAs and Their Targets in Platanus acerifolia under Lead (Pb) Stress
Abstract
:1. Introduction
2. Results and Discussion
2.1. Analyses of Small RNAs of P. acerifolia
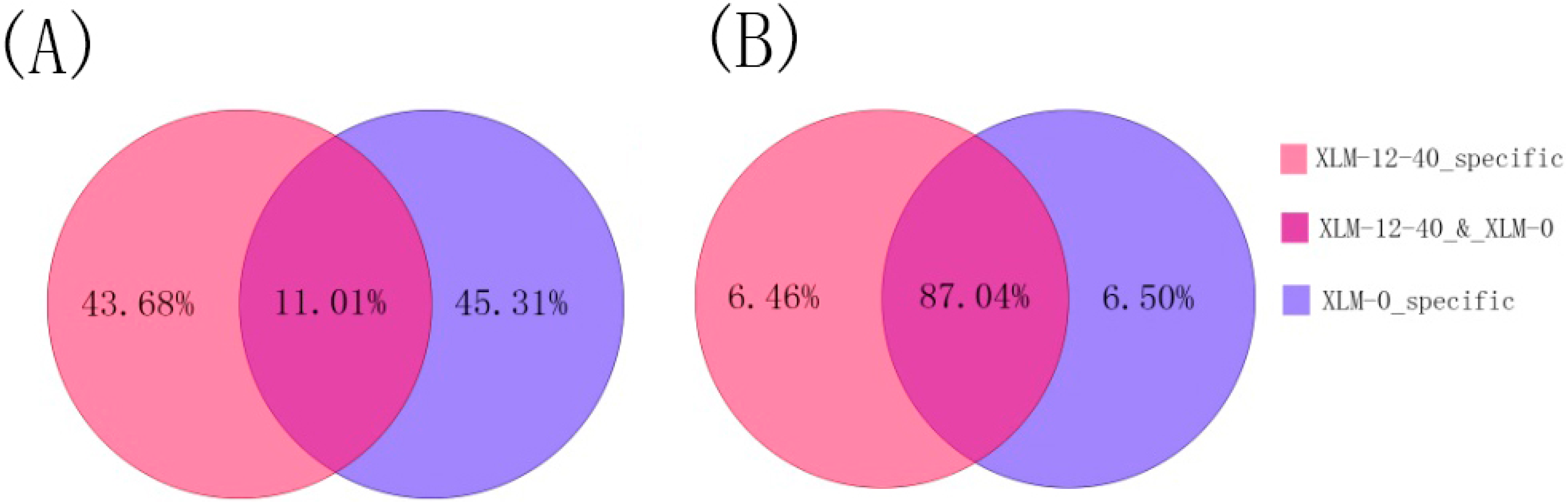
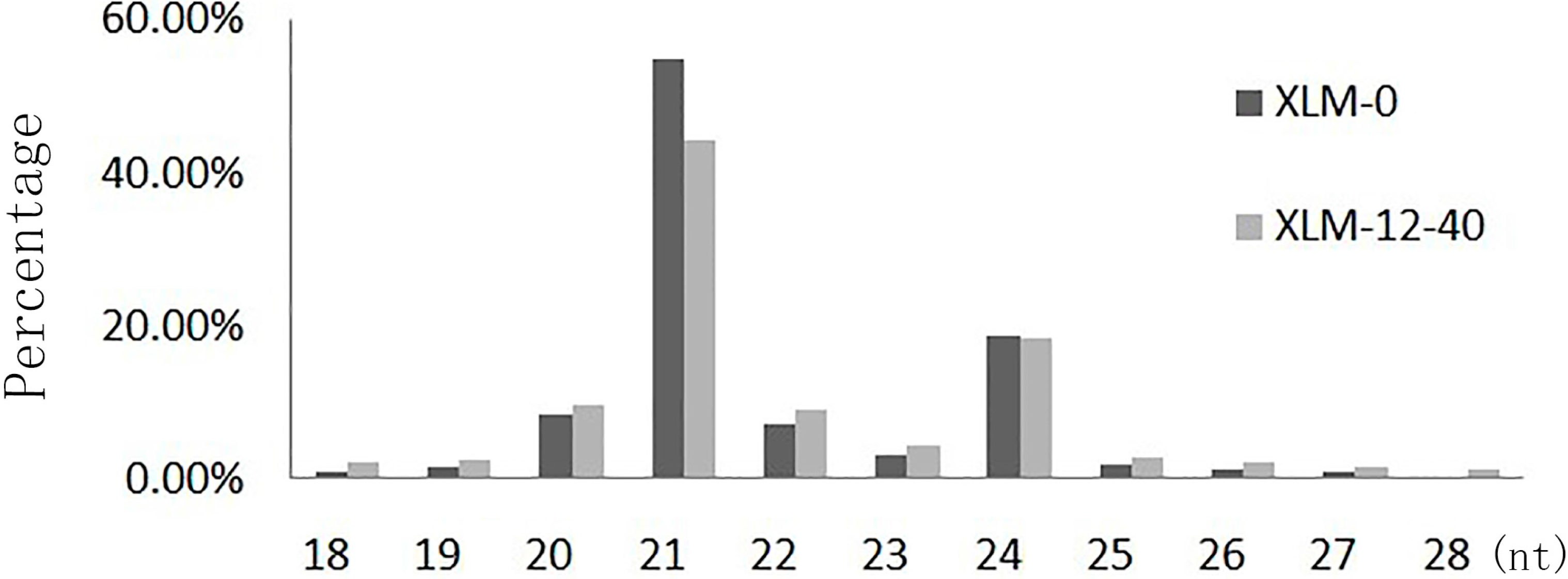
| Category | xlm-0 | xlm-12-40 | ||||||
|---|---|---|---|---|---|---|---|---|
| Unique sRNAs | Percent% | Total | Percent% | Unique | Percent% | Total | Percent% | |
| sRNAs | sRNAs | sRNAs | ||||||
| total | 1,938,169 | 100% | 13,993,838 | 100% | 1,882,090 | 100% | 13,818,458 | 100% |
| miRNA | 13,016 | 0.67% | 6,795,537 | 48.56% | 11,264 | 0.60% | 4,945,635 | 35.79% |
| rRNA | 105,837 | 5.46% | 1,544,175 | 11.03% | 126,468 | 6.72% | 2,464,243 | 17.83% |
| snRNA | 1206 | 0.06% | 3845 | 0.03% | 1816 | 0.10% | 10,279 | 0.07% |
| snoRNA | 483 | 0.02% | 1747 | 0.01% | 795 | 0.04% | 3989 | 0.03% |
| tRNA | 14,316 | 0.74% | 578,095 | 4.13% | 19,698 | 1.05% | 1,226,773 | 8.88% |
| unannote | 1,803,311 | 93.04% | 5,070,439 | 36.23% | 1,722,049 | 91.50% | 5,167,539 | 37.40% |
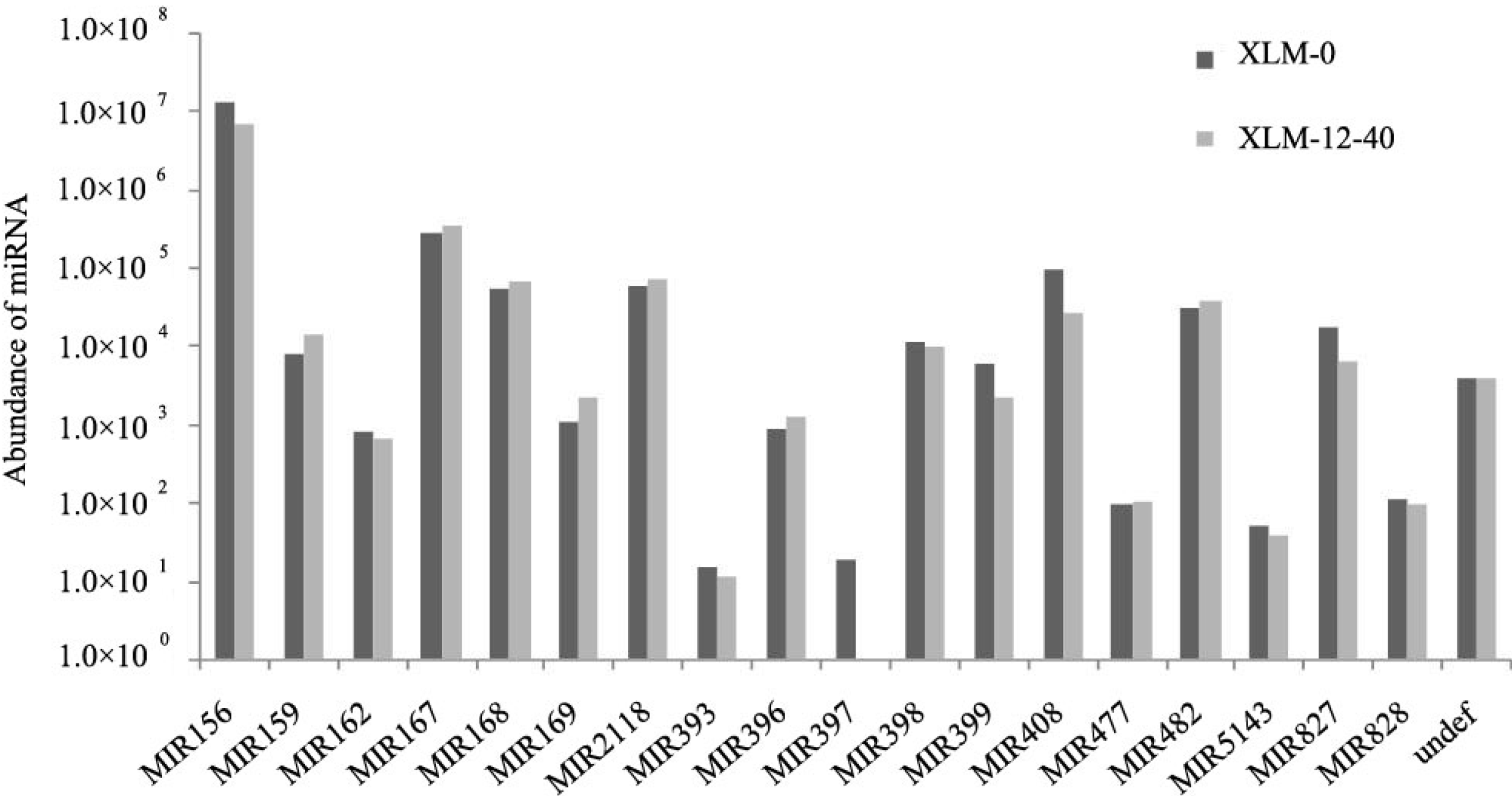
2.2. Identification of Conserved miRNAs in P. acerifolia
2.3. Identification of Novel miRNAs in P. acerifolia
2.4. Analysis of the Differential Expression miRNAs under Pb Stress
2.5. Identification of miRNA Targets by Degradome Sequencing
2.6. Validation of miRNAs and Their Targets with qRT-PCR
2.7. Discussion
3. Experimental Section
3.1. Experimental Materials
3.2. Construction and Sequencing of P. acerifolia sRNA
3.3. Bioinformatics Analysis sRNA and Identification miRNA of P. acerifolia
3.4. Analysis of the Differentially Expressed miRNAs under Pb Stress
3.5. Identification of miRNA Targets with Degradome Sequencing
3.6. miRNA Verification by qRT-PCR
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Slam, E.; Yang, X.; Li, T.; Liu, D.; Jin, X.; Meng, F. Effect of Pb toxicity on root morphology, physiology and ultrastructure in the two ecotypes of Elsholtzia argyi. J. Hazard. Mater. 2007, 147, 806–816. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Yin, L.; Zhang, Q.; Wang, W. Effect of Pb toxicity on leaf growth, antioxidant enzyme activities, and photosynthesis in cuttings and seedlings of Jatropha curcas L. Environ. Sci. Pollut. Res. Int. 2012, 19, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Kopittke, P.M.; Asher, C.J.; Kopittke, R.A.; Menzies, N.W. Toxic effects of Pb2+ on growth of cowpea (Vigna unguiculata). Environ. Pollut. 2007, 150, 280–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.; Liu, D. Effects of Pb2+ on root growth, cell division, and nucleolus of Zea mays L. Bull. Environ. Contam. Toxicol. 2000, 65, 786–793. [Google Scholar] [CrossRef] [PubMed]
- Reichheld, J.P.; Vernoux, T.; Lardon, F.; van, M.M.; Inze, D. Specific checkpoints regulate plant cell cycle progression in response to oxidative stress. Plant J. 1999, 17, 647–656. [Google Scholar] [CrossRef]
- Sengar, R.S.; Gautam, M.; Sengar, R.S.; Garg, S.K.; Sengar, K.; Chaudhary, R. Lead stress effects on physiobiochemical activities of higher plants. Rev. Environ. Contam. Toxicol. 2008, 196, 73–93. [Google Scholar] [PubMed]
- Matsouka, I.; Beri, D.; Chinou, I.; Haralampidis, K.; Spyropoulos, C.G. Metals and selenium induce medicarpin accumulation and excretion from the roots of fenugreek seedlings: A potential detoxification mechanism. Plant Soil 2011, 150, 280–287. [Google Scholar]
- Besnard, G.; Tagmount, A.; Baradat, P.; Vigouroux, A.; Berville, A. Molecular approach of genetic affinities between wild and ornamental Platanus. Euphytica 2002, 126, 401–412. [Google Scholar] [CrossRef]
- Chen, Y.M. Landscape Dendrology. China Forestry Press: Beijing, China, 2011. [Google Scholar]
- Fang, L.J.; Shi, J.S.; He, Z.X. A study on character variation of the P. occidentalis L. seed source at seedling stage. J. Nanjing For. Univ. 2003, 27, 44–46. [Google Scholar]
- Wang, A.X.; Fang, Y.M. Tissue distribution of 6 kinds of transport heavy metal pollutants in Platanus hispanica leaves and annual branches. Plant Res. 2011, 4, 478–488. [Google Scholar]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Brodersen, P.; Sakvarelidze-Achard, L.; Bruun-Rasmussen, M.; Dunoyer, P.; Yamamoto, Y.Y.; Sieburth, L.; Voinnet, O. Widespread translational inhibition by plant miRNAs and siRNAs. Science 2008, 320, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, Y.; Watanabe, Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 2004, 101, 12753–12758. [Google Scholar] [CrossRef] [PubMed]
- Baumberger, N.; Baulcombe, D.C. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 2005, 102, 11928–11933. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.S.; Xie, Q.; Fei, J.F.; Chua, N.H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell 2005, 17, 1376–1386. [Google Scholar] [CrossRef] [PubMed]
- Floyd, S.K.; Bowman, J.L. Gene regulation: Ancient microRNA target sequences in plants. Nature 2004, 428, 485–486. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, M.W.; Reinhart, B.J.; Lim, L.P.; Burge, C.B.; Bartel, B.; Bartel, D.P. Prediction of plant microRNA targets. Cell 2002, 110, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Chen, Y.Q. Insights into the mechanism of plant development: Interactions of miRNAs pathway with phytohormone response. Biochem. Biophys. Res. Commun. 2009, 384, 1–5. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Zhu, S.; Zhang, B. MicroRNA-target gene responses to lead-induced stress in cotton (Gossypium hirsutum L.). Funct. Integr. Genomics 2014, 14, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Srivastava, A.K.; Suprasanna, P.; D’Souza, S.F. Identification and profiling of arsenic stress-induced microRNAs in Brassica juncea. J. Exp. Bot. 2013, 64, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhang, H. Molecular identification and analysis of arsenite stress-responsive miRNAs in rice. J. Agric. Food Chem. 2012, 60, 6524–6536. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, Q.; Pan, X. MicroRNAs and their regulatory roles in animals and plants. J. Cell. Physiol. 2007, 210, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Addo-Quaye, C.; Eshoo, T.W.; Bartel, D.P.; Axtell, M.J. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008, 18, 758–762. [Google Scholar] [CrossRef] [PubMed]
- German, M.A.; Pillay, M.; Jeong, D.H.; Hetawal, A.; Luo, S.; Janardhanan, P.; Kannan, V.; Rymarquis, L.A.; Nobuta, K.; German, R.; et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008, 26, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Addo-Quaye, C.; Miller, W.; Axtell, M.J. CleaveLand: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 2009, 25, 130–131. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.L.; Zhao, Y.; McDonald, H.; Zeng, T.; Hirst, M.; et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008, 18, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.; Hu, Z.; Jiang, Q.; Zhang, H. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 2013, 82, 113–129. [Google Scholar] [CrossRef] [PubMed]
- Naya, L.; Paul, S.; Valdes-Lopez, O.; Mendoza-Soto, A.B.; Nova-Franco, B.; Sosa-Valencia, G.; Reyes, J.L.; Hernandez, G. Regulation of copper homeostasis and biotic interactions by microRNA 398b in common bean. PLoS ONE 2014, 9, e84416. [Google Scholar] [CrossRef] [PubMed]
- Fahlgren, N.; Jogdeo, S.; Kasschau, K.D.; Sullivan, C.M.; Chapman, E.J.; Laubinger, S.; Smith, L.M.; Dasenko, M.; Givan, S.A.; Weigel, D.; et al. MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 2010, 22, 1074–1089. [Google Scholar] [CrossRef] [PubMed]
- Hou, R.; Bao, Z.; Wang, S.; Su, H.; Li, Y.; Du, H.; Hu, J.; Wang, S.; Hu, X. Transcriptome sequencing and de novo analysis for Yesso scallop (Patinopecten yessoensis) using 454 GS FLX. PLoS ONE 2011, 6, e21560. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, J.; Chauhan, R.; Swarnkar, M.K.; Chahota, R.K.; Singh, A.K.; Shankar, R.; Yadav, S.K. Comprehensive transcriptomic study on horse gram (Macrotyloma uniflorum): De novo assembly, functional characterization and comparative analysis in relation to drought stress. BMC Genomics 2013, 14, 647. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.K.; Cao, Z.H.; Hao, Y.J. Overexpression of a R2R3 MYB gene MdSIMYB1 increases tolerance to multiple stresses in transgenic tobacco and apples. Physiol. Plant. 2014, 150, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.Q.; Pan, X.Y.; Yu, Y.M.; Wang, W.Y.; Zhang, F.; Ge, Y.Y.; Shen, X.L.; Shen, F.Q.; Liu, X.J. De novo characterization of the Anthurium transcriptome and analysis of its digital gene expression under cold stress. BMC Genomics 2013, 14, 827. [Google Scholar] [CrossRef] [PubMed]
- Palmer, C.M.; Hindt, M.N.; Schmidt, H.; Clemens, S.; Guerinot, M.L. MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet. 2013, 9, e1003953. [Google Scholar] [CrossRef] [PubMed]
- Akagi, T.; Ikegami, A.; Yonemori, K. DkMyb2 wound-induced transcription factor of persimmon (Diospyros kaki Thunb.), contributes to proanthocyanidin regulation. Planta 2010, 232, 1045–1059. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.B.; Eticha, D.; Albacete, A.; Rao, I.M.; Roitsch, T.; Horst, W.J. Physiological and molecular analysis of the interaction between aluminium toxicity and drought stress in common bean (Phaseolus vulgaris). J. Exp. Bot. 2012, 63, 3109–3125. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Zhang, J. Effect of abscisic acid on active oxygen species, antioxidative defence system and oxidative damage in leaves of maize seedlings. Plant Cell Physiol. 2001, 42, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Van der Biezen, E.A.; Jones, J.D. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci 1998, 23, 454–456. [Google Scholar]
- Belkhadir, Y.; Subramaniam, R.; Dangl, J.L. Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr. Opin. Plant Biol. 2004, 7, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Van der Biezen, E.A.; Jones, J.D.G. The NB-ARC domain: A novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 1998, 8, R226–R228. [Google Scholar]
- Shiu, S.H.; Bleecker, A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef] [PubMed]
- Luan, M.; Xu, M.; Lu, Y.; Zhang, Q.; Zhang, L.; Zhang, C.; Fan, Y.; Lang, Z.; Wang, L. Family-wide survey of miR169s and NF-YAs and their expression profiles response to abiotic stress in maize roots. PLoS ONE 2014, 9, e91369. [Google Scholar] [CrossRef] [PubMed]
- Radwan, O.; Liu, Y.; Clough, S.J. Transcriptional analysis of soybean root response to Fusarium virguliforme, the causal agent of sudden death syndrome. Mol. Plant Microbe Interact. 2011, 24, 958–972. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gao, S.; Zhou, X.; Chellappan, P.; Chen, Z.; Zhou, X.; Zhang, X.; Fromuth, N.; Coutino, G.; Coffey, M.; et al. Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol. Biol. 2011, 75, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Gielen, H.; Remans, T.; Vangronsveld, J.; Cuypers, A. MicroRNAs in metal stress: Specific roles or secondary responses. Int. J. Mol. Sci. 2012, 13, 15826–15847. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Foyer, C.H. Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef] [PubMed]
- May, M.J.; Vernoux, T.; Leaver, C.; van Montagu, M.; Inzé, D. Glutathione homeostasis in plants: Implications for environmental sensing and plant development. J. Exp. Bot. 1998, 49, 649–667. [Google Scholar]
- Adamis, P.D.; Gomes, D.S.; Pinto, M.L.; Panek, A.D.; Eleutherio, E.C. The role of glutathione transferases in cadmium stress. Toxicol. Lett. 2004, 154, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Marrs, K.A. The functions and regulation of glutatione S-transferases in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 127–158. [Google Scholar] [CrossRef] [PubMed]
- Grill, E.; Mishra, S.; Srivastava, S.; Tripathi, R.D. Role of phytochelatins in phytoremediation of heavy metals. In Environmental Bioremediation Technologies; Singh, S., Tripathi, R., Eds.; Springer: Berlin, Germany, 2007; pp. 101–146. [Google Scholar]
- Mishra, S.; Tripathi, R.D.; Srivastava, S.; Dwivedi, S.; Trivedi, P.K.; Dhankher, O.P.; Khare, A. Thiol metabolism play significant role during cadmium detoxification by Ceratophyllum demersum L. Bioresour. Technol. 2009, 100, 2155–2161. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.J. The integration of glutathione homeostasis and redox signaling. J. Plant Physiol. 2008, 165, 1390–1403. [Google Scholar] [CrossRef] [PubMed]
- Mackowiak, S.D. Identification of novel and known miRNAs in deep-sequencing data with miRDeep2. Curr. Protoc. Bioinform. 2011. [Google Scholar] [CrossRef]
- Meyers, B.C.; Axtell, M.J.; Bartel, B.; Bartel, D.P.; Baulcombe, D.; Bowman, J.L.; Cao, X.; Carrington, J.C.; Chen, X.; Green, P.J.; et al. Criteria for annotation of plant MicroRNAs. Plant Cell 2008, 20, 3186–3190. [Google Scholar] [CrossRef] [PubMed]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhao, Z.; Deng, M.; Liu, R.; Niu, S.; Fan, G. Identification and Functional Analysis of MicroRNAs and Their Targets in Platanus acerifolia under Lead (Pb) Stress. Int. J. Mol. Sci. 2015, 16, 7098-7111. https://doi.org/10.3390/ijms16047098
Wang Y, Zhao Z, Deng M, Liu R, Niu S, Fan G. Identification and Functional Analysis of MicroRNAs and Their Targets in Platanus acerifolia under Lead (Pb) Stress. International Journal of Molecular Sciences. 2015; 16(4):7098-7111. https://doi.org/10.3390/ijms16047098
Chicago/Turabian StyleWang, Yuanlong, Zhenli Zhao, Minjie Deng, Rongning Liu, Suyan Niu, and Guoqiang Fan. 2015. "Identification and Functional Analysis of MicroRNAs and Their Targets in Platanus acerifolia under Lead (Pb) Stress" International Journal of Molecular Sciences 16, no. 4: 7098-7111. https://doi.org/10.3390/ijms16047098
APA StyleWang, Y., Zhao, Z., Deng, M., Liu, R., Niu, S., & Fan, G. (2015). Identification and Functional Analysis of MicroRNAs and Their Targets in Platanus acerifolia under Lead (Pb) Stress. International Journal of Molecular Sciences, 16(4), 7098-7111. https://doi.org/10.3390/ijms16047098





