Positive Selection and Functional Divergence of R2R3-MYB Paralogous Genes Expressed in Inflorescence Buds of Scutellaria Species (Labiatae)
Abstract
:1. Introduction
| Species | Pollen | Growth Form | |||
|---|---|---|---|---|---|
| Size (μm) | Exine | Stem | Inflorescence | Petal and Corolla | |
| S. indica | (18–23) × (12–17) | Finely reticulate | Erect or procumbent at base | Terminal loose raceme | Geniculate at base, purple, pink or white |
| S. tashiroi | (18–22) × (14–17) | Loose reticulate to rugulate | Slender, procumbent, tufted | Axillary, seldom terminal raceme | Curve at base, dark purple |
| S. playfairii | (16–20) × (10–15) | Loose reticulate to rugulate | Erect, seldom tufted | Terminal loose raceme | Geniculate at base, whitish purple |
| S. taiwanensis | (23–30) × (16–20) | Irregular rugulate | Erect, often tufted | Terminal loose raceme | Geniculate at base, white with purple spot |
| Group | Function | Species | Reference |
|---|---|---|---|
| MYB2/7/11 | Shoot and axillary meristems formation | Arabidopsis thaliana (S14) a | [28] |
| Flavonoid regulation through GA metabolism; regulate PAL, C4H, CHS, CHI and UFGT | Scutellaria baicalensis | [27] | |
| MYB8 | Induction of anthocyanin accumulation | Arabidopsis thaliana (S6) a and Nicotiana tabacum | [29] |
| Sharing similar expression pattern with C4H and CHS after GA treatment | Scutellaria baicalensis | [27] | |
| MYB13/19 | Alternation of expression level of anthocyanin biosynthesis genes and pigment accumulation under cold stress | Arabidopsis thaliana (AtMYB3) | [12] |
| Brassica oleracea | [30] | ||
| Nicotiana tabacum | [31] | ||
| MYB15 | Regulation of serine/threonine protein phosphatases to enhance salt or drought tolerance | Arabidopsis thaliana (AtMYB20) | [32,33] |
| MYB16 | MicroRNA regulation and anther and pollen development | Arabidopsis thaliana (S18) a | [17] |
2. Results
2.1. Gene Annotation by Basic Local Alignment Search Tool (BLAST) Analyses
| Group Name | Phylogenetic Grouping | tBLASTx to Arabidopsis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stas | Spla | Sind | Stai | Sbai | Smon | Amborella | Arabidopsis | Solanum | Oryza | Accession Number | E-Value | |
| MYB16 | KP167623 | KP167610 | KP167603 | KP167617 | KF008651 | ATYL_2013395 | – | – | XM_004236340 | – | NM_111977 | 7 × 10−69 |
| (Stas_9284) | (Spla_28842) | (Sind_28842) | (Stai_7654) | (SbMYB16) | (AtMYB65) | |||||||
| MYB15 | KP167621 | KP167607 | KP167598 | – | KF008664 | ATYL_2028518 | XM_006837947 | NM_105294 | XM_004236642 | NM_001070300 | NM_105294 | 1 × 10−85 |
| (Stas_14132) | (Spla_13224) | (Sind_16195) | (AtMYB20) | NM_001054563 | (AtMYB20) | |||||||
| NM_121666 | NM_001063857 | |||||||||||
| (AtMYB43) | NM_001068382 | |||||||||||
| NM_001069653 | ||||||||||||
| MYB8 | KP167618 | KP167606 | KP167604 | KP167613 | KF008657 | ATYL_2121808 | XM_006849579 | AF048841 | XM_004252468 | AF062915 | 3 × 10−57 | |
| (AtMYB82) | (AtMYB90) | |||||||||||
| NM_123397 | ||||||||||||
| (AtMYB23) | – | |||||||||||
| MYB2/7/11 | XM_006837947 | XM_004245674 | NM_001186451 | |||||||||
| XM_004248305 | ||||||||||||
| MYB11 | KP167619 | KP167609 | KP167600 | – | KF008660 | ATYL_2012934 | – | – | – | – | NM_125143 | 7 × 10−63 |
| (SbMYB11) | (AtMYB36) | |||||||||||
| MYB2/7 | – | KP167611 | KP167605 | KP167615 | KC990835 | ATYL_2108188/2121208 | – | – | – | – | AF062901 | 9 × 10−71 |
| (SbMYB2) | (AtMYB68) | |||||||||||
| KC990836 | ||||||||||||
| (SbMYB7) | ||||||||||||
| MYB13/19 | XM_006854550 | - | XM_004253144 | |||||||||
| XM_004246040 | ||||||||||||
| XM_004244680 | ||||||||||||
| MYB13 | KP167622 | KP167608 | KP167601 | KP167616 | KF008662 | – | – | – | – | – | NM_112200 | 3 × 10−40 |
| KP167599 | (SbMYB13) | (AtMYB5) | ||||||||||
| MYB19 | KP167620 | KP167612 | KP167602 | KP167614 | KF008667 | ATYL_2029067 | – | – | – | – | NM_112200 | 1 × 10−61 |
| (SbMYB19) | (AtMYB5) | |||||||||||
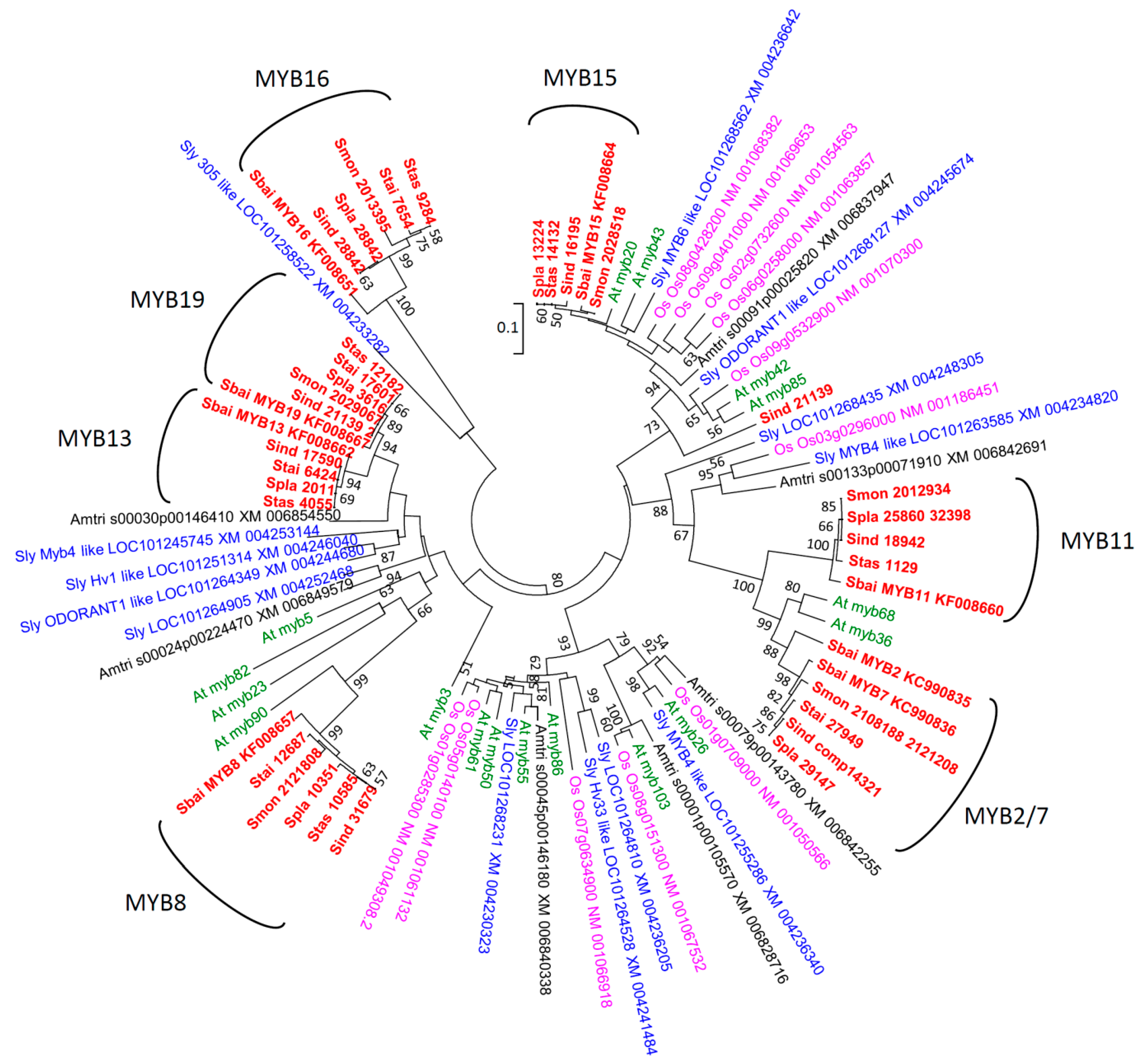
2.2. Phylogenetic Analyses
2.3. Codon-Specific Positive Selection

| Model | AtMYB5-Like | S6 | S14 | AtMYB20 | S18 | |||
|---|---|---|---|---|---|---|---|---|
| MYB 19 | MYB 13 | MYB 8 | MYB 7 | MYB 11 | MYB 15 | MYB 16 | ||
| M1a | lnL | −1869.28 | −1419.48 | −2050.95 | −1685.91 | −1763.79 | −1803.50 | −2587.30 |
| M2a | lnL | −1869.28 | −1419.24 | −2050.84 | −1685.59 | −1761.03 | −1802.53 | −2584.73 |
| 2ΔL | 0 | 0.49 | 0.22 | 0.63 | 5.52 | 1.94 | 5.13 | |
| p | 1 | 0.391 | 0.449 | 0.364 | 0.032 | 0.19 | 0.038 | |
| M7 | lnL | −1869.74 | −1419.61 | −2051.38 | −1686.13 | −1764.06 | −1804.13 | −2587.32 |
| M8 | lnL | −1869.28 | −1419.24 | −2050.84 | −1685.59 | −1762.24 | −1802.53 | −2584.74 |
| 2ΔL | 0.92 | 0.74 | 1.07 | 1.07 | 3.63 | 3.2 | 5.17 | |
| p | 0.491 | 0.345 | 0.293 | 0.293 | 0.082 | 0.101 | 0.038 | |
| M8a | lnL | −1869.28 | −1419.48 | −2050.95 | −1685.91 | −1763.80 | −1803.51 | −2587.34 |
| M8 | lnL | −1869.28 | −1419.24 | −2050.84 | −1685.59 | −1762.24 | −1802.53 | −2584.74 |
| 2ΔL | 0 | 0.49 | 0.22 | 0.63 | 3.11 | 1.96 | 5.19 | |
| p | 0.5 | 0.447 | 0.755 | 0.365 | 0.048 | 0.107 | 0.013 | |
2.4. Cluster-Specific Positive Selection and Functional Divergence
| Background | Amborella | Arabidopsis | Oryza | Solanum | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M1a (Null model) | ||||||||||||
| np | 101 | 113 | 109 | 121 | ||||||||
| lnL | −2402.594 | −2695.469 | −2735.734 | −3170.464 | ||||||||
| Site class | Class 0 | Class 1 | Class 0 | Class 1 | Class 0 | Class 1 | Class 0 | Class 1 | ||||
| Proportion | 0.938 | 0.062 | 0.869 | 0.131 | 0.824 | 0.176 | 0.99999 | 0.00001 | ||||
| ω | 0.042 | 1 | 0.041 | 1 | 0.042 | 1 | 0.039 | 1 | ||||
| Clade Model C | ||||||||||||
| np | 110 | 122 | 118 | 130 | ||||||||
| lnL | −2321.832 | −2606.658 | −2656.505 | −3059.730 | ||||||||
| Site class | Class 0 | Class 1 | Class 2 | Class 0 | Class 1 | Class 2 | Class 0 | Class 1 | Class 2 | Class 0 | Class 1 | Class 2 |
| Proportion | 0.698 | 0 | 0.302 | 0.710 | 0 | 0.290 | 0.676 | 0.034 | 0.290 | 0.687 | 0 | 0.313 |
| ωBackground | 0.010 | 1 | 0.154 | 0.013 | 1 | 0.183 | 0.014 | 1 | 0.285 | 0.010 | 1 | 0.134 |
| ωMYB2/7 | 0.010 | 1 | 0.062 | 0.013 | 1 | 0.101 | 0.014 | 1 | 0.040 | 0.010 | 1 | 0.045 |
| ωMYB11 | 0.010 | 1 | 0.037 | 0.013 | 1 | 0.196 | 0.014 | 1 | 0.097 | 0.010 | 1 | 0.056 |
| ωMYB15 | 0.010 | 1 | 0.028 | 0.013 | 1 | 0.000 | 0.014 | 1 | 0.017 | 0.010 | 1 | 0.000 |
| ωMYB16 | 0.010 | 1 | 0.207 | 0.013 | 1 | 0.114 | 0.014 | 1 | 0.085 | 0.010 | 1 | 0.039 |
| ωMYB8 | 0.010 | 1 | 0.054 | 0.013 | 1 | 0.058 | 0.014 | 1 | 0.029 | 0.010 | 1 | 0.160 |
| ωMYB13 | 0.010 | 1 | 0.000 | 0.013 | 1 | 0.000 | 0.014 | 1 | 0.000 | 0.010 | 1 | 0.000 |
| ωMYB19 | 0.010 | 1 | 999 | 0.013 | 1 | 999 | 0.014 | 1 | 999 | 0.010 | 1 | 999 |
| LRT | ||||||||||||
| 2ΔL | 161.525 | 177.623 | 158.457 | 221.469 | ||||||||
| df | 9 | 9 | 9 | 9 | ||||||||
| p | 3.58 × 10−30 | 1.59 × 10−33 | 1.55 × 10−29 | 1.03 × 10−42 | ||||||||
| Model | np | lnL | Parameter | Class 0 | Class 1 | Class 2a | Class 2b |
|---|---|---|---|---|---|---|---|
| Model A ω = 1 fixed | 82 | −1486.769 | Proportion | 0.7630 | 0.1728 | 0.0523 | 0.0119 |
| Background ω | 0.0285 | 1 | 0.0285 | 1 | |||
| Foreground ω | 0.0285 | 1 | 1 | 1 | |||
| Model A | 83 | −1484.552 | Proportion | 0.7930 | 0.1798 | 0.0222 | 0.0050 |
| Background ω | 0.0292 | 1 | 0.0292 | 1 | |||
| Foreground ω | 0.0292 | 1 | 11.1973 | 11.1973 | |||
| LRT a | 2ΔL = 4.434, p = 0.0176 | ||||||
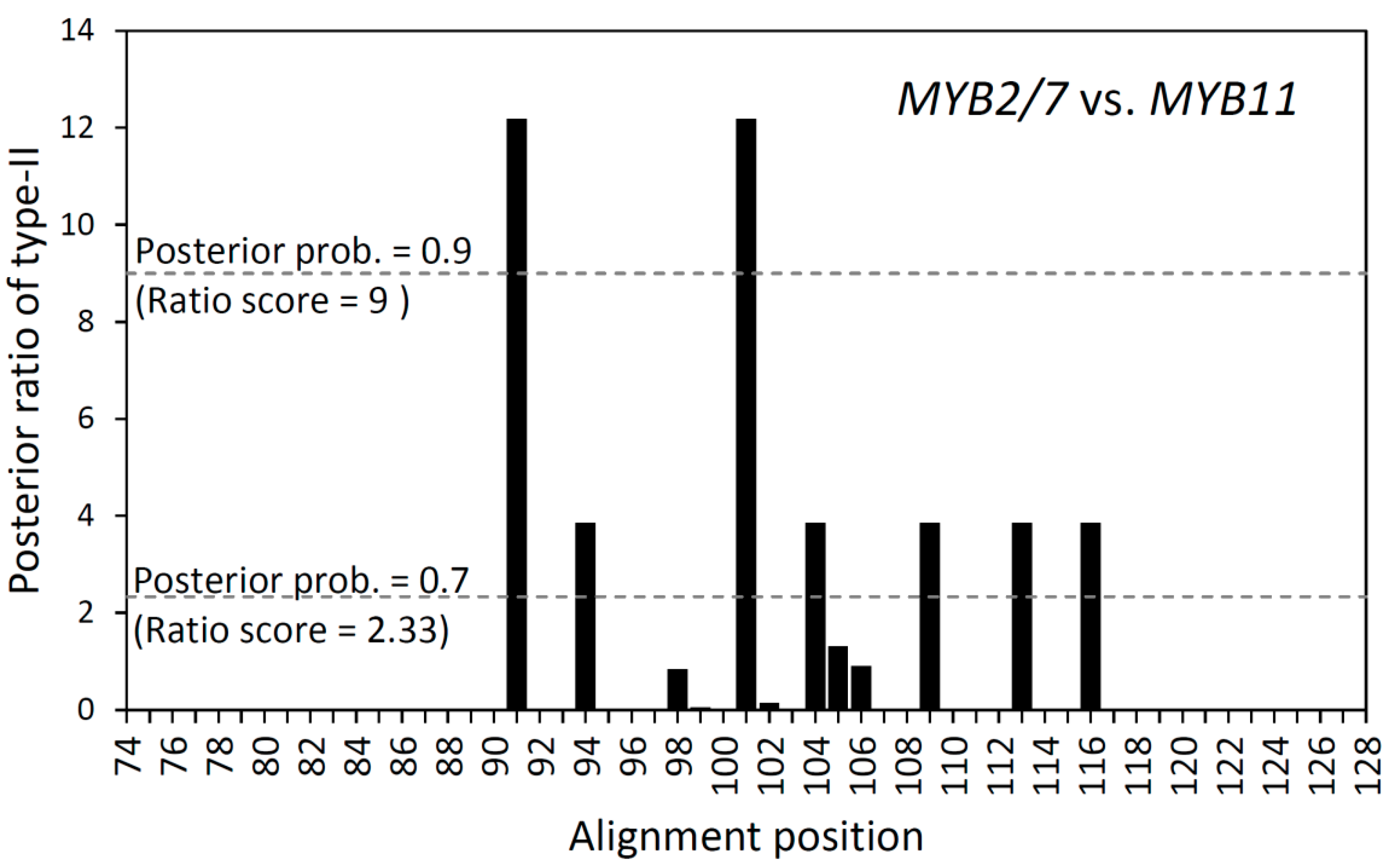
| Type-I Functional Divergence | Type-II Functional Divergence | ||||
|---|---|---|---|---|---|
| Parameter | MYB19 vs. MYB13 | MYB7/11 vs. MYB2 | Parameter | MYB19 vs. MYB13 | MYB7/11 vs. MYB2 |
| θI | 1.021 | −0.413 | θII | 0.052 | 0.125 |
| SE θI | 0.161 | 0.293 | SE θII | 0.036 | 0.072 |
| p of θI Z-score | <0.00001 | 0.099 | p of θII Z-score | 0.073 | 0.042 |
| θIML | 0.999 | 0.006 | aR/πR | 1.405 | 1.869 |
| AlphaML | 0.006 | 0.126 | GR/GC | 1 | 0.864 |
| SE θI | 0.234 | 0.050 | F00,N | 0.927 | 0.727 |
| LRT θI | 18.289 | 0.003 | F00,R | 0.018 | 0.036 |
| p of LRT θI | 1.898 × 10−5 | 0.956 | F00,C | 0.018 | 0.091 |
3. Discussion
3.1. Diversification of R2R3-MYBs in Scutellaria Expressed in Inflorescence Buds
3.2. Phylogenetic Analyses and Functional Annotation
3.3. Positive Selection on Scutellaria MYB11 and MYB16
3.4. Different Types of Functional Divergence between Recent Duplicated Paralogs in Scutellaria R2R3-MYBs
4. Experimental Section
4.1. Data Collection, de Novo Transcriptome Assembly, and Annotation
4.2. Detecting Positive Selection
4.3. Functional Divergence Analyses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mol, J.; Grotewold, E.; Koes, R. How genes paint flowers and seeds. Trends Plant Sci. 1998, 3, 212–217. [Google Scholar] [CrossRef]
- Malcomber, S.T.; Preston, J.C.; Reinheimer, R.; Kossuth, J.; Kellogg, E.A. Developmental gene evolution and the origin of grass inflorescence diversity. Adv. Bot. Res. 2006, 44, 425–481. [Google Scholar]
- Linhart, Y.B.; Grant, M.C. Evolutionary significance of local genetic differentiation in plants. Annu. Rev. Ecol. Syst. 1996, 27, 237–277. [Google Scholar] [CrossRef]
- Evans, L.M.; Slavov, G.T.; Rodgers-Melnick, E.; Martin, J.; Ranjan, P.; Muchero, W.; Brunner, A.M.; Schackwitz, W.; Gunter, L.; Chen, J.G.; et al. Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations. Nat. Genet. 2014, 46, 1089–1096. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.A.; Porter, A.H. Rapid speciation via parallel, directional selection on regulatory genetic pathways. J. Theor. Biol. 2000, 205, 527–542. [Google Scholar] [CrossRef] [PubMed]
- Rebollo, R.; Horard, B.; Hubert, B.; Vieira, C. Jumping genes and epigenetics: Towards new species. Gene 2010, 454, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.B.W.; Lindell, J.; Backstrom, N. Speciation genetics: Current status and evolving approaches. Philos. Trans. B 2010, 365, 1717–1733. [Google Scholar] [CrossRef]
- Huang, T.-C. Notes on the Flora of Taiwan (35)-Scutellaria taipeiensis T.C. Huang, A. Hsiao et M.J. Wu sp. nov. (Lamiaceae). Taiwania 2003, 48, 129–137. [Google Scholar]
- Chiang, Y.C.; Huang, B.H.; Liao, P.C. Diversification, biogeographic pattern, and demographic history of Taiwanese Scutellaria species inferred from nuclear and chloroplast DNA. PLoS One 2012, 7, e50844. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.T.H.; Linthorst, H.J.M.; Verpoorte, R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011, 10, 397–412. [Google Scholar] [CrossRef] [PubMed]
- Tohge, T.; Nishiyama, Y.; Hirai, M.Y.; Yano, M.; Nakajima, J.; Awazuhara, M.; Inoue, E.; Takahashi, H.; Goodenowe, D.B.; Kitayama, M.; et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005, 42, 218–235. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Keller, T.; Abbott, J.; Moritz, T.; Doerner, P. Arabidopsis regulator of axillary meristems1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell Online 2006, 18, 598–611. [Google Scholar] [CrossRef]
- Müller, D.; Schmitz, G.; Theres, K. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell Online 2006, 18, 586–597. [Google Scholar] [CrossRef]
- Baumann, K.; Perez-Rodriguez, M.; Bradley, D.; Venail, J.; Bailey, P.; Jin, H.L.; Koes, R.; Roberts, K.; Martin, C. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 2007, 134, 1691–1701. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are MicroRNA-regulated genes that redundantly facilitate anther development. Plant Cell 2005, 17, 705–721. [Google Scholar] [CrossRef] [PubMed]
- Fraser, L.G.; Seal, A.G.; Montefiori, M.; McGhie, T.K.; Tsang, G.K.; Datson, P.M.; Hilario, E.; Marsh, H.E.; Dunn, J.K.; Hellens, R.P.; et al. An R2R3 MYB transcription factor determines red petal colour in an Actinidia (kiwifruit) hybrid population. BMC Genomics 2013, 14, 28. [Google Scholar] [CrossRef] [PubMed]
- Streisfeld, M.A.; Young, W.N.; Sobel, J.M. Divergent selection drives genetic differentiation in an R2R3-MYB transcription factor that contributes to incipient speciation in Mimulus aurantiacus. PLoS Genet. 2013, 9, e1003385. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, R.; Yamagishi, N.; Yoshikawa, N. A MYB transcription factor controls flower color in soybean. J. Hered. 2013, 104, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.N.; Mao, Y.; Liu, H.L.; Yu, F.X.; Li, S.X.; Yin, T.M. Transcriptome analysis of differentially expressed genes relevant to variegation in peach flowers. PLoS One 2014, 9, e90842. [Google Scholar] [CrossRef] [PubMed]
- Uematsu, C.; Katayama, H.; Makino, I.; Inagaki, A.; Arakawa, O.; Martin, C. Peace, a MYB-like transcription factor, regulates petal pigmentation in flowering peach “Genpei” bearing variegated and fully pigmented flowers. J. Exp. Bot. 2014, 65, 1081–1094. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, R.; Rausher, M.D. Pollinator-mediated selection on flower color allele drives reinforcement. Science 2012, 335, 1090–1092. [Google Scholar] [CrossRef] [PubMed]
- Liao, P.C.; Huang, B.H. National Taiwan Normal University: Taiwan, Unpublished work. 2015.
- Dias, A.P.; Braun, E.L.; McMullen, M.D.; Grotewold, E. Recently duplicated maize R2R3 Myb genes provide evidence for distinct mechanisms of evolutionary divergence after duplication. Plant Physiol. 2003, 131, 610–620. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E.; Sainz, M.B.; Tagliani, L.; Hernandez, J.M.; Bowen, B.; Chandler, V.L. Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl. Acad. Sci. USA 2000, 97, 13579–13584. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Wu, C.; Liu, Y.J.; Yang, J.; Huang, L.Q. The Scutellaria baicalensis R2R3-MYB transcription factors modulates flavonoid biosynthesis by regulating GA metabolism in transgenic tobacco plants. PLoS One 2013, 8, e77275. [Google Scholar] [CrossRef] [PubMed]
- Kranz, H.D.; Denekamp, M.; Greco, R.; Jin, H.; Leyva, A.; Meissner, R.C.; Petroni, K.; Urzainqui, A.; Bevan, M.; Martin, C. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 1998, 16, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.A.; Lamb, C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell Online 2000, 12, 2383–2393. [Google Scholar] [CrossRef]
- Zhang, B.; Hu, Z.; Zhang, Y.; Li, Y.; Zhou, S.; Chen, G. A putative functional MYB transcription factor induced by low temperature regulates anthocyanin biosynthesis in purple kale (Brassica oleracea var. acephala f. tricolor). Plant Cell Rep. 2012, 31, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.A.; Zhao, T.; Fan, H.J.; Wang, N.; Zheng, S.S.; Ling, H.Q. The upregulation of NtAN2 expression at low temperature is required for anthocyanin accumulation in juvenile leaves of Lc-transgenic Tobacco (Nicotiana tabacum L.). J. Genet. Genomics 2012, 39, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.H.; Yoo, K.S.; Hyoung, S.; Nguyen, H.T.K.; Kim, Y.Y.; Kim, H.J.; Ok, S.H.; Yoo, S.D.; Shin, J.S. An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 2013, 587, 1773–1778. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zhang, Y.L.; Yang, L.; Song, J.B.; Yang, Z.M. AtMYB20 is negatively involved in plant adaptive response to drought stress. Plant Soil 2014, 376, 433–443. [Google Scholar] [CrossRef]
- Dubos, C.; le Gourrierec, J.; Baudry, A.; Huep, G.; Lanet, E.; Debeaujon, I.; Routaboul, J.M.; Alboresi, A.; Weisshaar, B.; Lepiniec, L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008, 55, 940–953. [Google Scholar] [CrossRef] [PubMed]
- Ballerini, E.S.; Mockaitis, K.; Arnold, M.L. Transcriptome sequencing and phylogenetic analysis of floral and leaf MIKCC MADS-box and R2R3 MYB transcription factors from the monocot Iris fulva. Gene 2013, 531, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Bhalla, P.L.; Singh, M.B. Transcriptome-wide profiling and expression analysis of transcription factor families in a liverwort, Marchantia polymorpha. BMC Genomics 2013, 14, 915. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.L.; Tang, Y.; Zhang, K.X.; Li, F.L.; Yang, P.Y.; Tang, Y.X.; Wu, Y.M.; Shao, J.R. Identification of TT2 gene from floral transcriptome in Fagopyrum tataricum. Food Res. Int. 2013, 54, 1331–1333. [Google Scholar] [CrossRef]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.J.; Campbell, M.M. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009, 149, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Cole, I.B.; Cao, J.; Alan, A.R.; Saxena, P.K.; Murch, S.J. Comparisons of Scutellaria baicalensis, Scutellaria lateriflora and Scutellaria racemosa: Genome size, antioxidant potential and phytochemistry. Planta Medica 2008, 74, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Qi, L.; Yang, J.; Wu, C.; Liu, Y.; Huang, L. A Scutellaria baicalensis R2R3-MYB gene, SbMYB8, regulates flavonoid biosynthesis and improves drought stress tolerance in transgenic tobacco. Plant Cell Tissue Organ Cult. 2014, 1–12. [Google Scholar]
- Hsieh, K.; Huang, A.H.C. Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell 2007, 19, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Stroo, A. Pollen morphological evolution in bat pollinated plants. Plant Syst. Evol. 2000, 22, 225–242. [Google Scholar] [CrossRef]
- Jamzad, Z.; Hasani-Nejad, M. Taxonomic implications of pollen exine morphology in infrageneric classification of Scutellaria (Lamiaceae). Nord. J. Bot. 2014, 32, 233–244. [Google Scholar] [CrossRef]
- Chang, S.M.; Lu, Y.Q.; Rausher, M.D. Neutral evolution of the nonbinding region of the anthocyanin regulatory gene Ipmyb1 in Ipomoea. Genetics 2005, 170, 1967–1978. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Clegg, M.T.; Jiang, T. Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol. 2004, 134, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.M.; Heine, G.F.; Irani, N.G.; Feller, A.; Kim, M.G.; Matulnik, T.; Chandler, V.L.; Grotewold, E. Different mechanisms participate in the R-dependent activity of the R2R3 MYB transcription factor C1. J. Biol. Chem. 2004, 279, 48205–48213. [Google Scholar] [CrossRef] [PubMed]
- Peter, C.I.; Johnson, S.D. A pollinator shift explains floral divergence in an orchid species complex in South Africa. Ann. Bot. 2014, 113, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Nowak, M.A.; Boerlijst, M.C.; Cooke, J.; Smith, J.M. Evolution of genetic redundancy. Nature 1997, 388, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A. Genetic redundancy caused by gene duplications and its evolution in networks of transcriptional regulators. Biol. Cybern. 1996, 74, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Z. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298. [Google Scholar] [CrossRef]
- Bergthorsson, U.; Andersson, D.I.; Roth, J.R. Ohno’s dilemma: Evolution of new genes under continuous selection. Proc. Natl. Acad. Sci. USA 2007, 104, 17004–17009. [Google Scholar] [CrossRef] [PubMed]
- Nougue, O.; Corbi, J.; Ball, S.G.; Manicacci, D.; Tenaillon, M.I. Molecular evolution accompanying functional divergence of duplicated genes along the plant starch biosynthesis pathway. BMC Evolut. Biol. 2014, 14, 103. [Google Scholar] [CrossRef]
- Osborn, T.C.; Pires, J.C.; Birchler, J.A.; Auger, D.L.; Chen, Z.J.; Lee, H.S.; Comai, L.; Madlung, A.; Doerge, R.W.; Colot, V.; et al. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 2003, 19, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Wolfe, K.H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 2004, 16, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Renny-Byfield, S.; Gallagher, J.P.; Grover, C.E.; Szadkowski, E.; Page, J.T.; Udall, J.A.; Wang, X.Y.; Paterson, A.H.; Wendel, J.F. Ancient gene duplicates in Gossypium (Cotton) exhibit near-complete expression divergence. Genome Biol. Evolut. 2014, 6, 559–571. [Google Scholar] [CrossRef]
- Guo, H.; Lee, T.H.; Wang, X.Y.; Paterson, A.H. Function relaxation followed by diversifying selection after whole-genome duplication in flowering plants. Plant Physiol. 2013, 162, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.A.; Wang, Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 2011, 12, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.H. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Zou, Y.Y.; Su, Z.X.; Huang, W.; Zhou, Z.; Arendsee, Z.; Zeng, Y.W. An update of DIVERGE software for functional divergence analysis of protein family. Mol. Biol. Evol. 2013, 30, 1713–1719. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Xu, D.P.; Gu, X. Functional divergence after gene duplication and sequence-structure relationship: A case study of G-protein α subunits. J. Exp. Zool. 2007, 308B, 85–96. [Google Scholar] [CrossRef]
- Streisfeld, M.A.; Liu, D.; Rausher, M.D. Predictable patterns of constraint among anthocyanin-regulating transcription factors in Ipomoea. New Phytol. 2011, 191, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Streisfeld, M.A.; Rausher, M.D. Relaxed constraint and evolutionary rate variation between basic helix-loop-helix floral anthocyanin regulators in Ipomoea. Mol. Biol. Evol. 2007, 24, 2816–2826. [Google Scholar] [CrossRef] [PubMed]
- Carroll, S.B. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 2008, 134, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Doebley, J.; Lukens, L. Transcriptional regulators and the evolution of plant form. Plant Cell 1998, 10, 1075–1082. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, B.-H.; Pang, E.; Chen, Y.-W.; Cao, H.; Ruan, Y.; Liao, P.-C. Positive Selection and Functional Divergence of R2R3-MYB Paralogous Genes Expressed in Inflorescence Buds of Scutellaria Species (Labiatae). Int. J. Mol. Sci. 2015, 16, 5900-5921. https://doi.org/10.3390/ijms16035900
Huang B-H, Pang E, Chen Y-W, Cao H, Ruan Y, Liao P-C. Positive Selection and Functional Divergence of R2R3-MYB Paralogous Genes Expressed in Inflorescence Buds of Scutellaria Species (Labiatae). International Journal of Molecular Sciences. 2015; 16(3):5900-5921. https://doi.org/10.3390/ijms16035900
Chicago/Turabian StyleHuang, Bing-Hong, Erli Pang, Yi-Wen Chen, Huifen Cao, Yu Ruan, and Pei-Chun Liao. 2015. "Positive Selection and Functional Divergence of R2R3-MYB Paralogous Genes Expressed in Inflorescence Buds of Scutellaria Species (Labiatae)" International Journal of Molecular Sciences 16, no. 3: 5900-5921. https://doi.org/10.3390/ijms16035900





