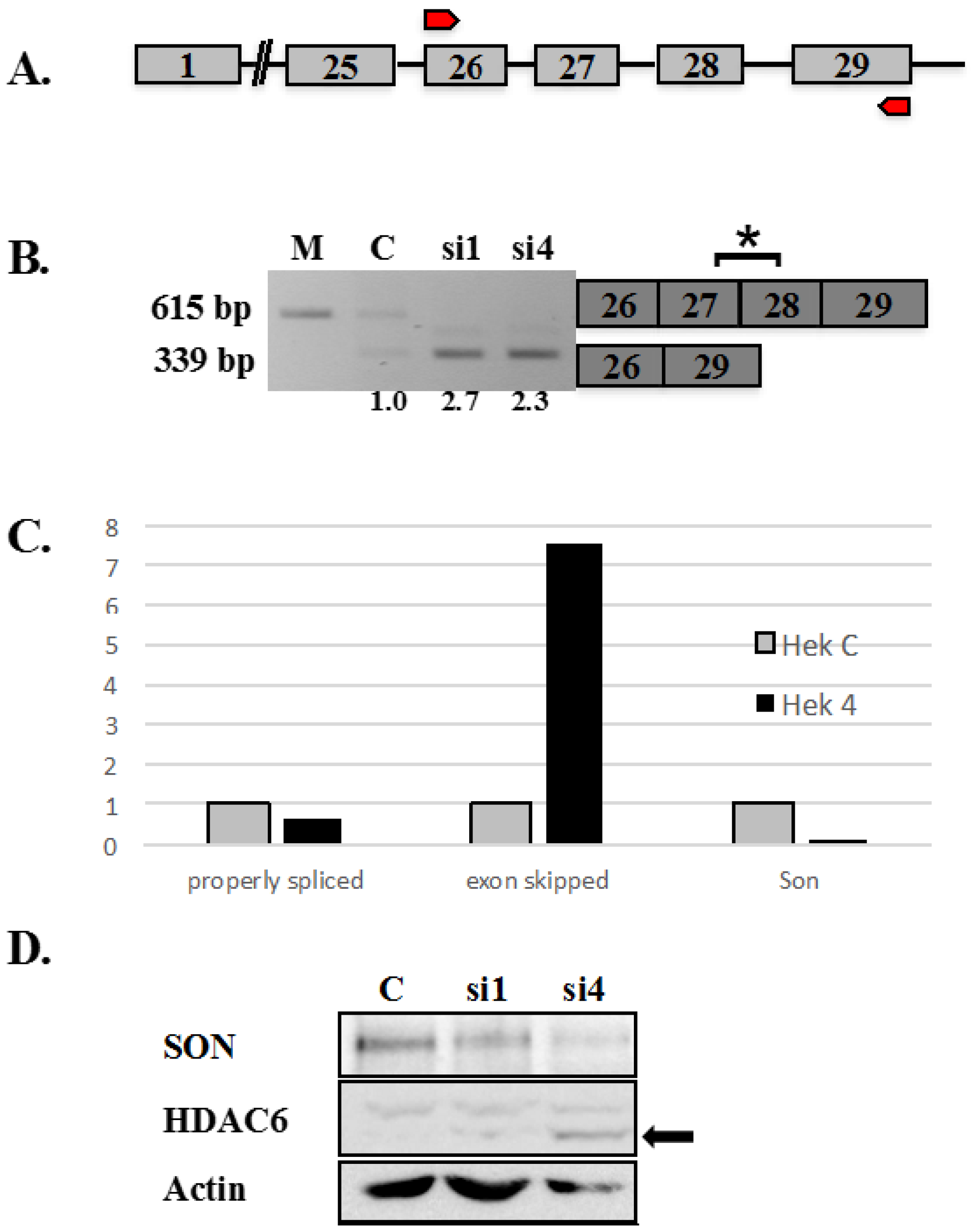
2.1. SON Is Required for Retention of Exons 27 and 28 in Endogenous HDAC6 Transcripts and Exogenous HDAC6 Minigene Reporter Transcripts
Reducing SON expression level affected all different types of splicing, including exon skipping [
9,
10,
11]. SON depletion resulted in skipping of exon 6 in rat beta-tropomyosin (BTM) minigene reporter transcripts. Misregulated splicing was also shown in a subset of human transcripts including skipping of exons 27 and 28 in endogenous HDAC6 transcripts ([
9] and
Figure 1B). Histone deacetylases (HDACs) remove acetyl groups from nucleosomal histones at conserved lysine residues, as a critical step of controlling chromatin accessibility and transcription activity during gene regulation. HDACs can also deacetylate lysine residues of transcription factors and thereby regulate their DNA binding ability. HDAC6 is a class IIB histone deacetylase and is a cytoplasmic protein (reviewed in [
12]). HDAC6 is the only histone deacetylase that contains two functional catalytic (CAT) domains and a
C-terminal BUZ (binding-of-ubiquitin zinc finger) domain [
13]. The two catalytic histone deacetylase domains function in deacetylating α-tubulin, playing a key role in cell motility [
14]. The HDAC6 BUZ-domain (amino acids 1134–1192) is encoded by 174 nucleotides contained within exons 27 and 28 of HDAC6 mRNA.
We tested whether SON depletion that results in skipping exons 27 and 28 in HDAC transcripts would ultimately interfere with BUZ domain functions. Gene structure of HDAC6 is shown in
Figure 1A, indicating exons as numbered boxes, connected by lines that represent introns. Transcripts including sequence spanning from the beginning of exon 26 through the end of exon 29 (indicated by red arrows in
Figure 1A) are the focus of our current study. As shown previously, exons 27 and 28 of endogenous HDAC6 transcripts were skipped in SON-depleted cells but not in cells treated with control siRNAs (
Figure 1B and [
9]). Importantly, we also detected skipping of exons 27 and 28 in HDAC6 transcripts in human embryonic kidney cells (
Figure 1C). Our studies did not determine whether altered splicing of HDAC6 transcripts affects HDAC6 protein expression or HDAC6 function. To test this, we examined the effects of SON depletion on HDAC6 protein. Immunoblot data in
Figure 1 demonstrates that endogenous HDAC6 protein has a molecular mass roughly 10 kDa smaller in extracts from SON-depleted HeLa cells compared to HDAC6 in extracts from controls (
Figure 1D); this truncated protein is predicted to not contain the BUZ domain due to absence of exons 27 and 28 in the mature HDAC6 mRNA under these conditions. This result suggests a biological relevance for SON for maintaining HDAC6 functions in human cells. We next compared patterns of exon skipping for endogenous
versus reporter HDAC6 transcripts. Notably, incorrectly spliced HDAC6 transcripts were evident in samples having partial SON reduction, indicating that reduced SON expression is adequate for disrupting splicing of HDAC6 transcripts.
Figure 1.
Altered splicing of HDAC6 pre-mRNA in SON depleted HeLa cells. (
A) Human HDAC6 contains 29 exons represented as boxes and the intervening introns represented as lines joining exons. Red arrows represent the region of genomic DNA used to construct HDAC6 minigene reporter plasmid shown in
Figure 2,
Figure 3 and
Figure 4; (
B) RT-PCR performed using primers that amplify endogenous HDAC6 transcripts shows exon skipping in SON siRNA-treated samples compared to mock (M) and control (C). Boxes to the right indicate the region of HDAC6 transcripts detected by RT-PCR. Asterisk indicates exons 27 and 28 are skipped in SON-depleted cells. Numerical values below lanes represent the ratio of transcripts having retained:excluded introns; (
C) Human embryonic kidney cells treated with either control siRNAs or si4 siRNA duplex targeting SON (Hek 4) were used for qRT-PCR with HDAC6 exon-junction oligos. SON depleted cells showed significantly reduced expression of SON transcripts (Son) as well as reduction of properly spliced HDAC6 transcripts, but showed increased skipping of exons 27 and 28 (exon skipped) for endogenous HDAC6 transcripts; and (
D) HeLa cells were transfected with control siRNA oligo (C) and two different siRNA oligos targeting SON. Forty-eight hours post-transfection, protein was extracted and immunoblotting was performed with antibodies against SON (WU09), HDAC6 and actin. Western blot confirmed reduced SON protein in cells treated with siRNA duplexes targeting SON. Arrow indicates that the molecular mass of HDAC6 is 10 kDa smaller following treatment with siRNA targeting SON compared to control siRNA treated cells.
Figure 1.
Altered splicing of HDAC6 pre-mRNA in SON depleted HeLa cells. (
A) Human HDAC6 contains 29 exons represented as boxes and the intervening introns represented as lines joining exons. Red arrows represent the region of genomic DNA used to construct HDAC6 minigene reporter plasmid shown in
Figure 2,
Figure 3 and
Figure 4; (
B) RT-PCR performed using primers that amplify endogenous HDAC6 transcripts shows exon skipping in SON siRNA-treated samples compared to mock (M) and control (C). Boxes to the right indicate the region of HDAC6 transcripts detected by RT-PCR. Asterisk indicates exons 27 and 28 are skipped in SON-depleted cells. Numerical values below lanes represent the ratio of transcripts having retained:excluded introns; (
C) Human embryonic kidney cells treated with either control siRNAs or si4 siRNA duplex targeting SON (Hek 4) were used for qRT-PCR with HDAC6 exon-junction oligos. SON depleted cells showed significantly reduced expression of SON transcripts (Son) as well as reduction of properly spliced HDAC6 transcripts, but showed increased skipping of exons 27 and 28 (exon skipped) for endogenous HDAC6 transcripts; and (
D) HeLa cells were transfected with control siRNA oligo (C) and two different siRNA oligos targeting SON. Forty-eight hours post-transfection, protein was extracted and immunoblotting was performed with antibodies against SON (WU09), HDAC6 and actin. Western blot confirmed reduced SON protein in cells treated with siRNA duplexes targeting SON. Arrow indicates that the molecular mass of HDAC6 is 10 kDa smaller following treatment with siRNA targeting SON compared to control siRNA treated cells.
![Ijms 16 05886 g001]()
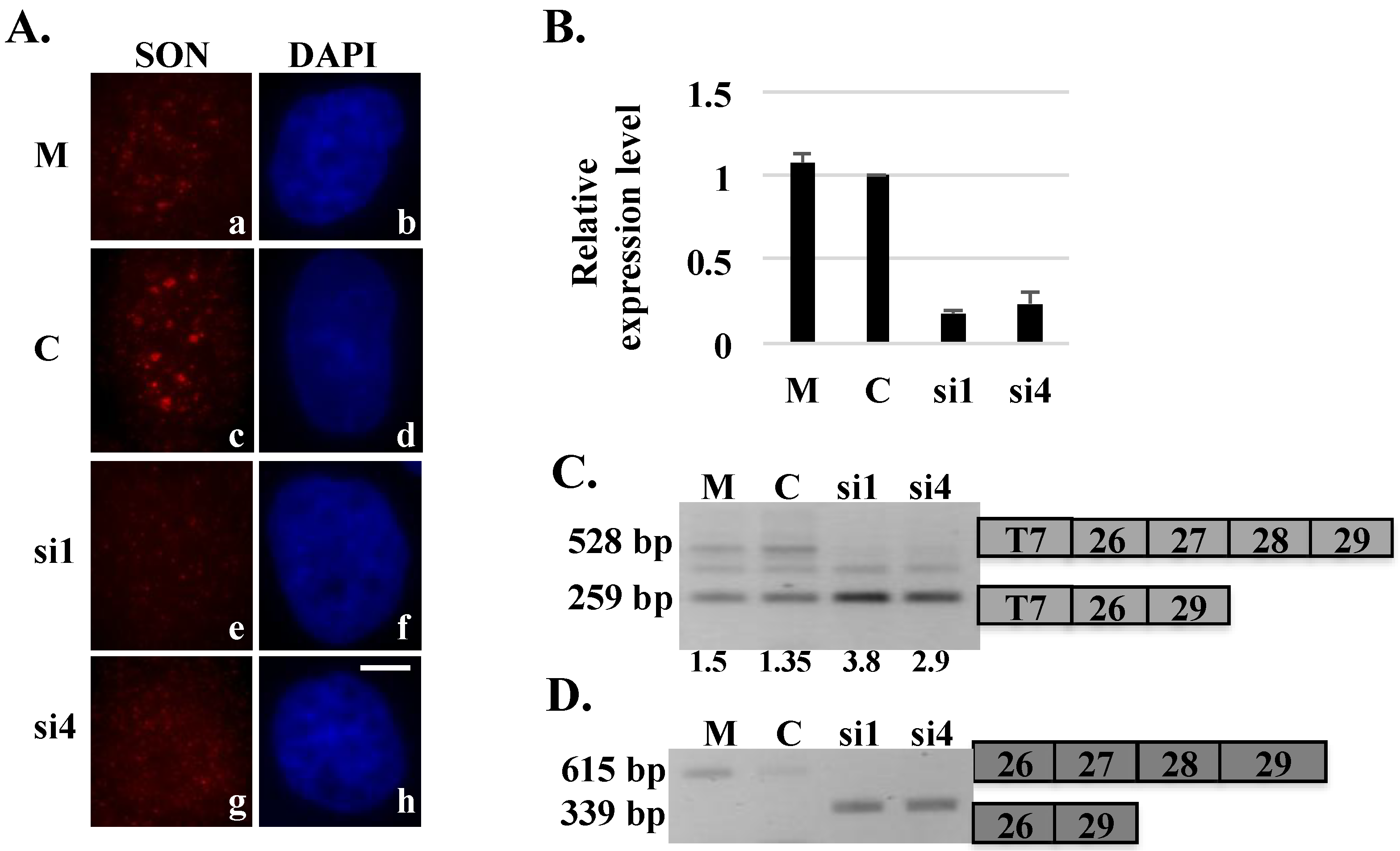
Reporter minigenes typically contain a segment of genomic DNA corresponding to regions of a gene that become alternatively spliced (or mis-spliced) under a specific condition. When expressed in cells, these minigene reporters allow in
in vivo analysis of
cis-regulatory elements (such as cryptic splice sites, exonic splicing enhancers and exonic splicing silencers) and
trans-acting factors such as splicing factors and splicing repressors [
15]. To examine splicing changes in exogenous HDAC6 transcripts upon SON depletion, HeLa cells transiently transfected with plasmid expressing our human HDAC6 reporter minigene, were treated with siRNA oligos targeting SON mRNA. Reduction of SON transcripts was confirmed by qRT-PCR (
Figure 2B). SON-depleted cells showed reduced immunolabeling of SON in nuclear speckles compared control siRNA-treated cells (
Figure 2A; Sharma
et al., 2010 [
9]). Polyacrylamide gel electrophoresis of RT-PCR products indicated that properly spliced HDAC6 transcripts containing both exons 27 and 28 were detected in control cells, while exons 27 and 28 were predominantly skipped in HDAC6 transcripts from SON-depleted cells, and properly spliced transcripts were not detected (
Figure 2C).
Figure 2.
Skipping of exons 27 and 28 of the HDAC6 reporter minigene in SON-depleted cells. HeLa cells transiently transfected with HDAC6 reporter plasmid and treated with mock (M), control (C) or SON siRNA duplexes were either processed for immunofluorescence (A) or harvested to perform qRT-PCR (B) to show SON depletion. Cells treated with SON siRNAs and immunolabeled with anti-SON antibodies in (A) do not show SON labeling in nuclear speckles (e and g) when compared to mock treated (a) or control siRNA-treated (c) cells. DNA was stained with DAPI (b, d, f and h). Scale bar = 5 µm. qRT-PCR in (B) showed reduction of SON mRNA levels in SON siRNA-treated samples compared to mock and control siRNA treated samples; (C) RT-PCR performed using primers that specifically target reporter minigene transcripts shows exon skipping in SON siRNA-treated samples.
Figure 2.
Skipping of exons 27 and 28 of the HDAC6 reporter minigene in SON-depleted cells. HeLa cells transiently transfected with HDAC6 reporter plasmid and treated with mock (M), control (C) or SON siRNA duplexes were either processed for immunofluorescence (A) or harvested to perform qRT-PCR (B) to show SON depletion. Cells treated with SON siRNAs and immunolabeled with anti-SON antibodies in (A) do not show SON labeling in nuclear speckles (e and g) when compared to mock treated (a) or control siRNA-treated (c) cells. DNA was stained with DAPI (b, d, f and h). Scale bar = 5 µm. qRT-PCR in (B) showed reduction of SON mRNA levels in SON siRNA-treated samples compared to mock and control siRNA treated samples; (C) RT-PCR performed using primers that specifically target reporter minigene transcripts shows exon skipping in SON siRNA-treated samples.
To further investigate SON-mediated splicing of HDAC6 transcripts, we constructed HeLa cell lines stably expressing our HDAC6 reporter minigene. Three HeLa HDAC6 clones stably expressing the HDAC6 reporter minigene were transfected with SON siRNA to test splicing patterns of HDAC6 minigene reporter transcripts. SON depletion was performed in three separate HeLa HDAC6 stable cell lines that each consistently showed skipping of exons 27 and 28 both in exogenous HDAC6 transcripts (
Figure 3C, shows one representative data set for clone 69) and in endogenous HDAC6 transcripts (
Figure 3D). The rationale for testing multiple clones was based upon our hypothesis that independent clones that presumably contain HDAC6 minigene integrated randomly at different genomic locations may yield different splicing results if chromatin context influences splicing of HDAC6 transcripts. However, the similarity of results among multiple independent clones indicates that chromatin context probably does not influence splicing of these specific exons.
Figure 3.
Stably expressed HDAC6 reporter transcripts show exon skipping after SON depletion. HeLa HDAC6 clone 69 stably expressing HDAC6 reporter plasmid was transfected with mock (M), control (C) or SON siRNA duplexes and processed for either immunofluorescence (A) or harvested to perform qRT-PCR (B) to show SON depletion. Cells treated with SON siRNA and immunolabeled with anti-SON antibodies in (A) do not show SON labeling in nuclear speckles (e and g) when compared to mock (a) or control siRNA (c) treated cells. DNA was stained with DAPI (b, d, f and h). Scale bar = 5 µm. qRT-PCR in (B) showed reduction of SON mRNA levels in SON siRNA-treated samples when compared to mock and control siRNA treated samples; (C) RT-PCR performed using primers that specifically target exogenous reporter HDAC6 minigene transcripts shows exon skipping in SON siRNA-treated samples; and (D) RT-PCR performed with primers that specifically target endogenous HDAC6 transcripts shows exon skipping in SON siRNA-treated samples. Numerical values below lanes in (C) represent the ratio of transcripts having retained:excluded introns.
Figure 3.
Stably expressed HDAC6 reporter transcripts show exon skipping after SON depletion. HeLa HDAC6 clone 69 stably expressing HDAC6 reporter plasmid was transfected with mock (M), control (C) or SON siRNA duplexes and processed for either immunofluorescence (A) or harvested to perform qRT-PCR (B) to show SON depletion. Cells treated with SON siRNA and immunolabeled with anti-SON antibodies in (A) do not show SON labeling in nuclear speckles (e and g) when compared to mock (a) or control siRNA (c) treated cells. DNA was stained with DAPI (b, d, f and h). Scale bar = 5 µm. qRT-PCR in (B) showed reduction of SON mRNA levels in SON siRNA-treated samples when compared to mock and control siRNA treated samples; (C) RT-PCR performed using primers that specifically target exogenous reporter HDAC6 minigene transcripts shows exon skipping in SON siRNA-treated samples; and (D) RT-PCR performed with primers that specifically target endogenous HDAC6 transcripts shows exon skipping in SON siRNA-treated samples. Numerical values below lanes in (C) represent the ratio of transcripts having retained:excluded introns.
![Ijms 16 05886 g003]()
2.2. Aberrant Splicing of HDAC6 in Absence of SON Is Biologically Significant
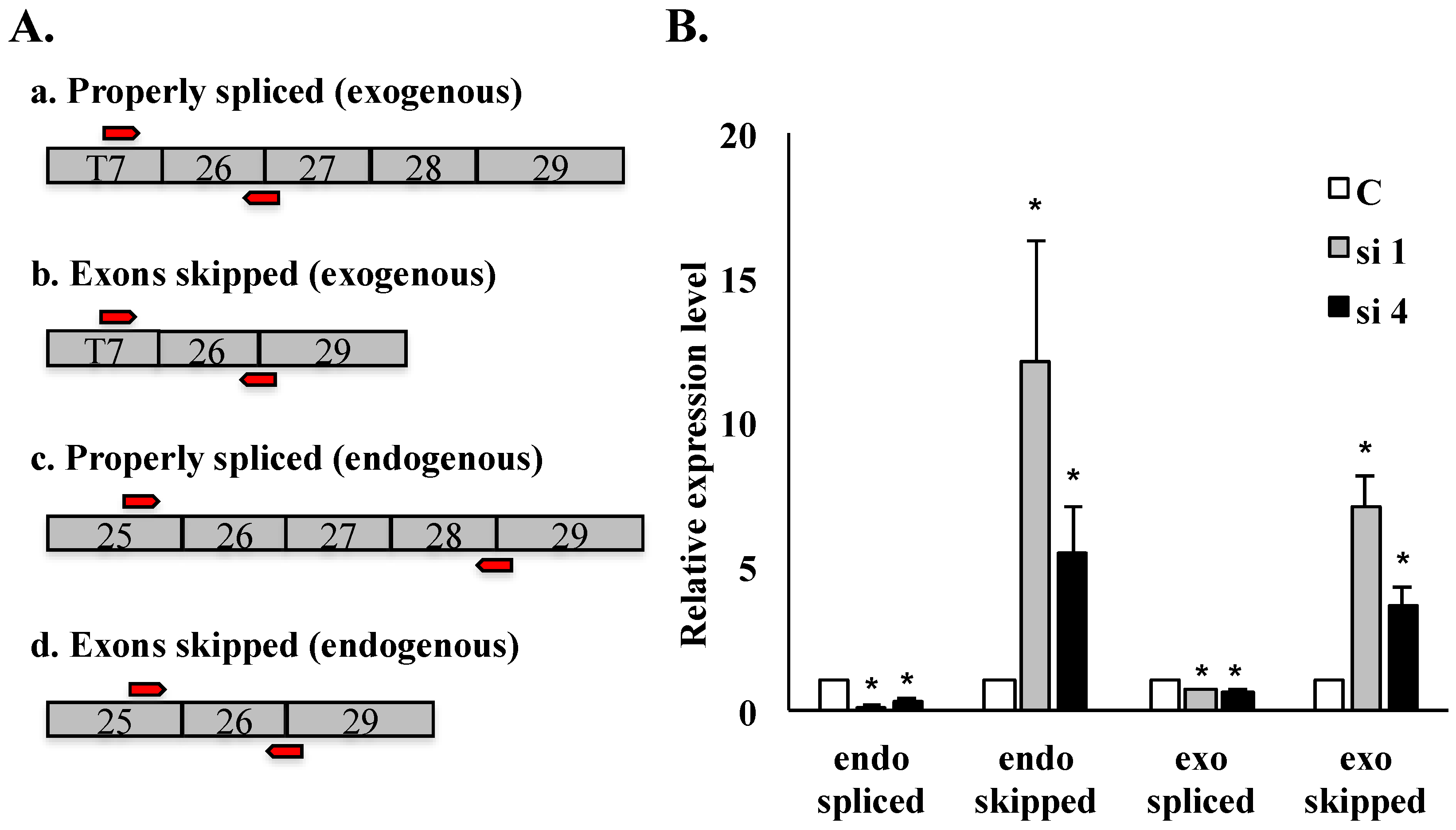
We performed quantitative RT-PCR to examine the abundance of properly spliced
versus mis-spliced HDAC6 transcripts in SON-depleted cells compared to controls. Primers spanning exon junctions specifically amplified properly spliced
versus mis-spliced transcripts, while primers targeted specifically to sequences upstream of exon 26 allowed detection of exogenous or endogenous HDAC6 transcripts as shown in
Figure 4A. qRT-PCR was performed in the SON-depleted HeLa HDAC6 stable cell line “clone 69”, and SON depletion was validated by qRT-PCR. In SON-depleted cells, the level of properly spliced HDAC6 transcripts was reduced (for both endogenous and exogenous HDAC6 transcripts) while the level of mis-spliced transcripts increased in comparison with respective transcript levels in cells treated with control siRNA (
Figure 4B). This data shows that mis-splicing of HDAC6 transcripts is more prevalent in SON-depleted cells, and it indicates the efficacy of our HDAC6 minigene reporter system for mimicking misregulated HDAC6 pre-mRNA splicing patterns.
Figure 4.
qRT-PCR analysis of HDAC6 exon skipping in SON-depleted cells. qRT-PCR performed on HeLa HDAC6 stable cells post-SON depletion. (A) Four sets of exon junction primers indicated by red arrows (see Experimental Section) were used to amplify splice variants of exogenous reporter transcripts as well as of endogenous HDAC6 transcripts; (B) Relative transcript level for properly spliced versus exon skipped transcripts is measured both for endogenous and for exogenous HDAC6 transcripts. Error bars were calculated by measuring standard error from three individual experiments (* p ≤ 0.05).
Figure 4.
qRT-PCR analysis of HDAC6 exon skipping in SON-depleted cells. qRT-PCR performed on HeLa HDAC6 stable cells post-SON depletion. (A) Four sets of exon junction primers indicated by red arrows (see Experimental Section) were used to amplify splice variants of exogenous reporter transcripts as well as of endogenous HDAC6 transcripts; (B) Relative transcript level for properly spliced versus exon skipped transcripts is measured both for endogenous and for exogenous HDAC6 transcripts. Error bars were calculated by measuring standard error from three individual experiments (* p ≤ 0.05).
Next, we performed
in situ analysis to test whether biological function of HDAC6 was disrupted in SON-depleted cells. An aggresome assembly assay determined that HDAC6 protein expressed in SON-depleted cells lacks normal BUZ domain function. Under normal cellular conditions, the HDAC6 BUZ domain associates with polyubiquitin and thus with ubiquitinated misfolded proteins [
16]. Formation of misfolded proteins increases under stress, and this condition can become toxic [
17]. In several human disorders, the accumulation of misfolded proteins leads to cell death. One mechanism of misfolded protein degradation involves HDAC6-dependent formation of aggresomes [
16]. HDAC6 interacts with dynein motor proteins as well as ubiquitinated misfolded proteins and directs them via microtubules to the microtubule-organizing center (MTOC), where aggresomes assemble [
16] and misfolded proteins are degraded by local proteases and autophagy proteins. Mutant HDAC6 lacking either of the catalytic deacetylase domains or the BUZ domain fails to support aggresome assembly, demonstrating the importance of these HDAC6 domains in aggresome function [
16].
To analyze HDAC6 function in SON depleted cells, we exogenously expressed GFP-CFTR ∆F508 (a straightforward approach to produce fluorescently labeled misfolded protein in cells) and then induced aggresome formation
in situ. These cells were then transfected with SON siRNA, and allowed time for SON depletion before aggresome formation was induced. Cells treated with control siRNA showed robust signal for GFP-CFTR ∆F508 in large clusters of cytoplasmic aggresomes (
Figure 5a,d, arrow), while diffuse cytoplasmic GFP signal was very low (
Figure 5a, filled arrowheads mark cytosol). In cells treated with SON siRNA, misfolded protein was predominantly cytoplasmic and diffuse (marked by filled arrowheads in
Figure 5e). An occasional SON-depleted cell showed small foci of GFP-CFTR ∆F508 (indicated by an asterisk in
Figure 5e) that could be explained by the low amount of properly spliced transcript detected in SON depleted cells that would produce “wild type” HDAC6 that may contribute to some minimal level of aggresome assembly. This is not unexpected, since the overall goal of this experiment was to see that in the majority of SON-depleted cells, the GFP-CFTR ∆F508 signal should be more dispersed due to the expression of truncated HDAC6 protein. Our results suggest that HDAC6 protein lacking the BUZ domain reduces the extent of aggresome assembly. Moreover, our results are consistent with data reported by Kawaguchi
et al., (2003) [
16] in that the HDAC6 protein lacking BUZ domain fails to support aggresome assembly.
Figure 5.
SON depletion reduces HDAC6-mediated aggresome formation. HeLa cells transiently expressing GFP-CFTR ∆F508 were transfected with either control siRNA duplexes (C) or SON siRNA duplexes and then treated with proteasome inhibitor MG132. Immunofluorescence labeling with anti-SON WU14 antibody indicates that SON normally localizes to nuclear speckles (open arrowheads in b) and that SON labeling is reduced in nuclear speckles of cells treated with SON siRNA duplexes (f). DAPI was used to stain DNA (c and g) and used to highlight the nuclear boundary in all panels (dotted lines). Arrow in (a–d) indicates GFP-CFTR ∆F508 localized at aggresomes. Asterisk in (e) marks a SON-depleted cell that retains a few cytoplasmic foci of GFP-CFTR ∆F508 (see Discussion). Filled arrowheads mark GFP-CFTR signal that is more diffuse in the cytoplasm of SON-depleted cells (e) while it is predominantly concentrated at aggresomes in controls (a). Triple merge shown in (d) and (h). Scale bar = 5 µm.
Figure 5.
SON depletion reduces HDAC6-mediated aggresome formation. HeLa cells transiently expressing GFP-CFTR ∆F508 were transfected with either control siRNA duplexes (C) or SON siRNA duplexes and then treated with proteasome inhibitor MG132. Immunofluorescence labeling with anti-SON WU14 antibody indicates that SON normally localizes to nuclear speckles (open arrowheads in b) and that SON labeling is reduced in nuclear speckles of cells treated with SON siRNA duplexes (f). DAPI was used to stain DNA (c and g) and used to highlight the nuclear boundary in all panels (dotted lines). Arrow in (a–d) indicates GFP-CFTR ∆F508 localized at aggresomes. Asterisk in (e) marks a SON-depleted cell that retains a few cytoplasmic foci of GFP-CFTR ∆F508 (see Discussion). Filled arrowheads mark GFP-CFTR signal that is more diffuse in the cytoplasm of SON-depleted cells (e) while it is predominantly concentrated at aggresomes in controls (a). Triple merge shown in (d) and (h). Scale bar = 5 µm.