Structural Analysis of the Complex between Penta-EF-Hand ALG-2 Protein and Sec31A Peptide Reveals a Novel Target Recognition Mechanism of ALG-2
Abstract
:1. Introduction
2. Results
2.1. Co-Crystallization
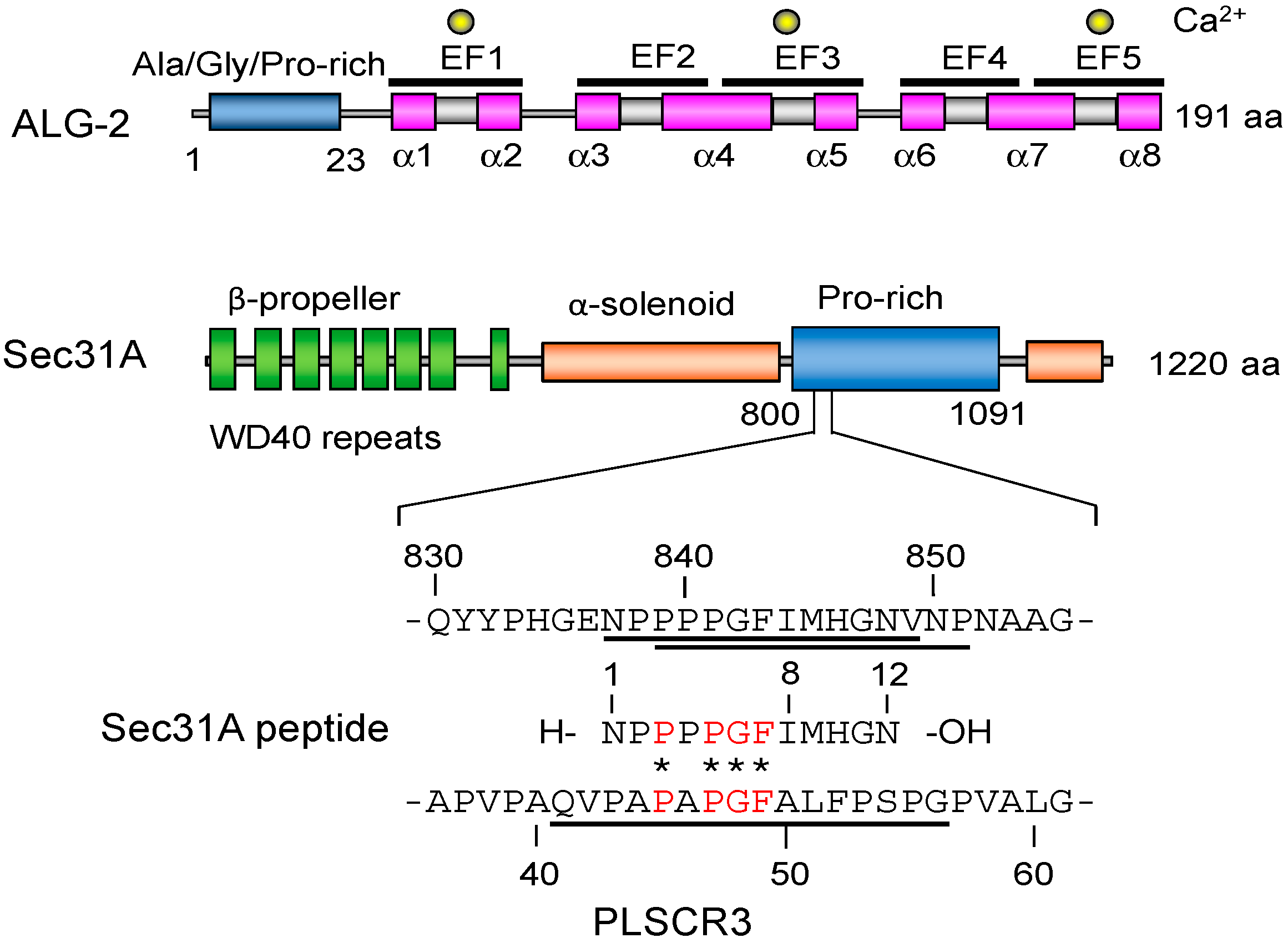
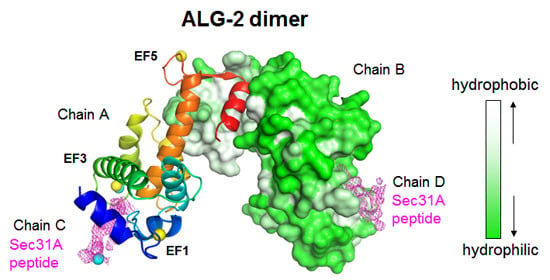
2.2. Overall Structure of the Complex between ALG-2 and Sec31A Peptide
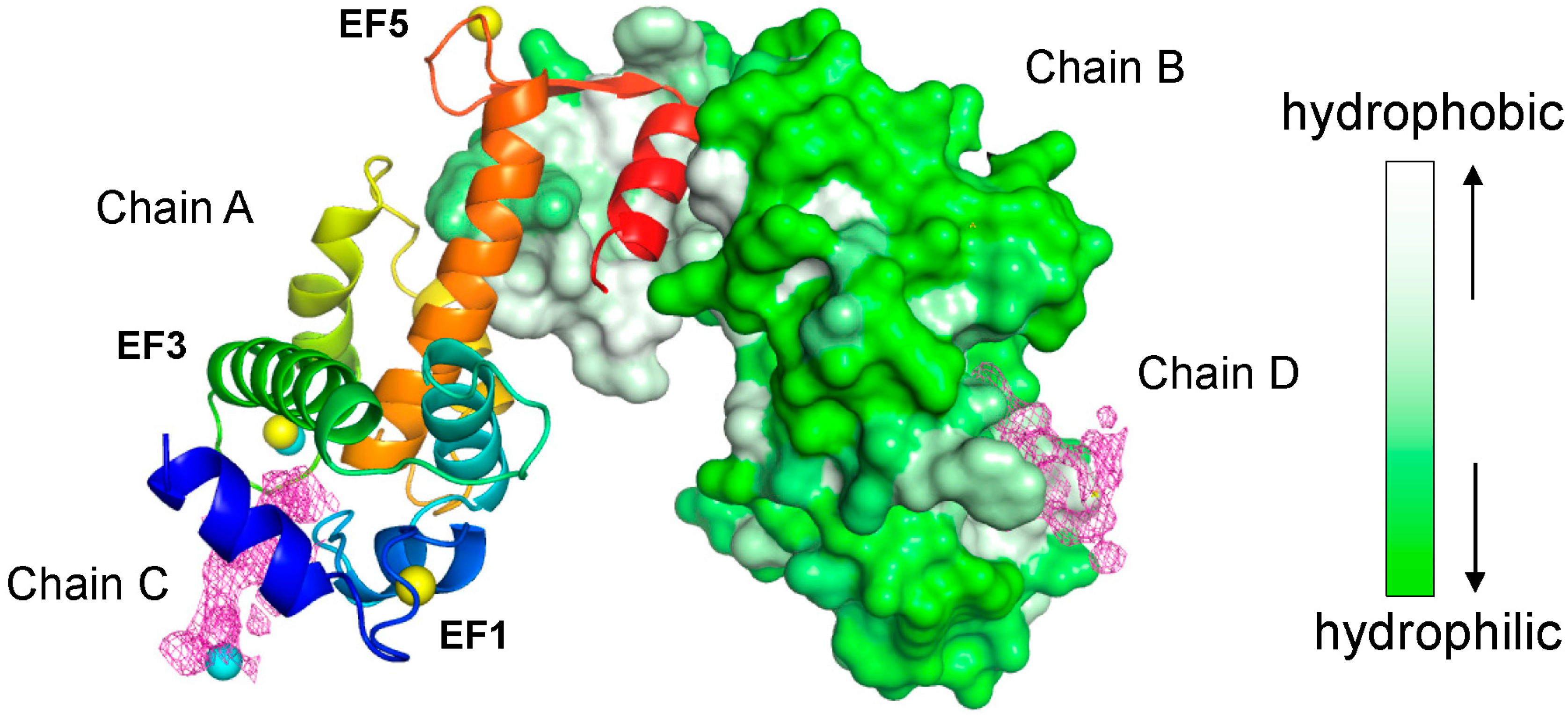
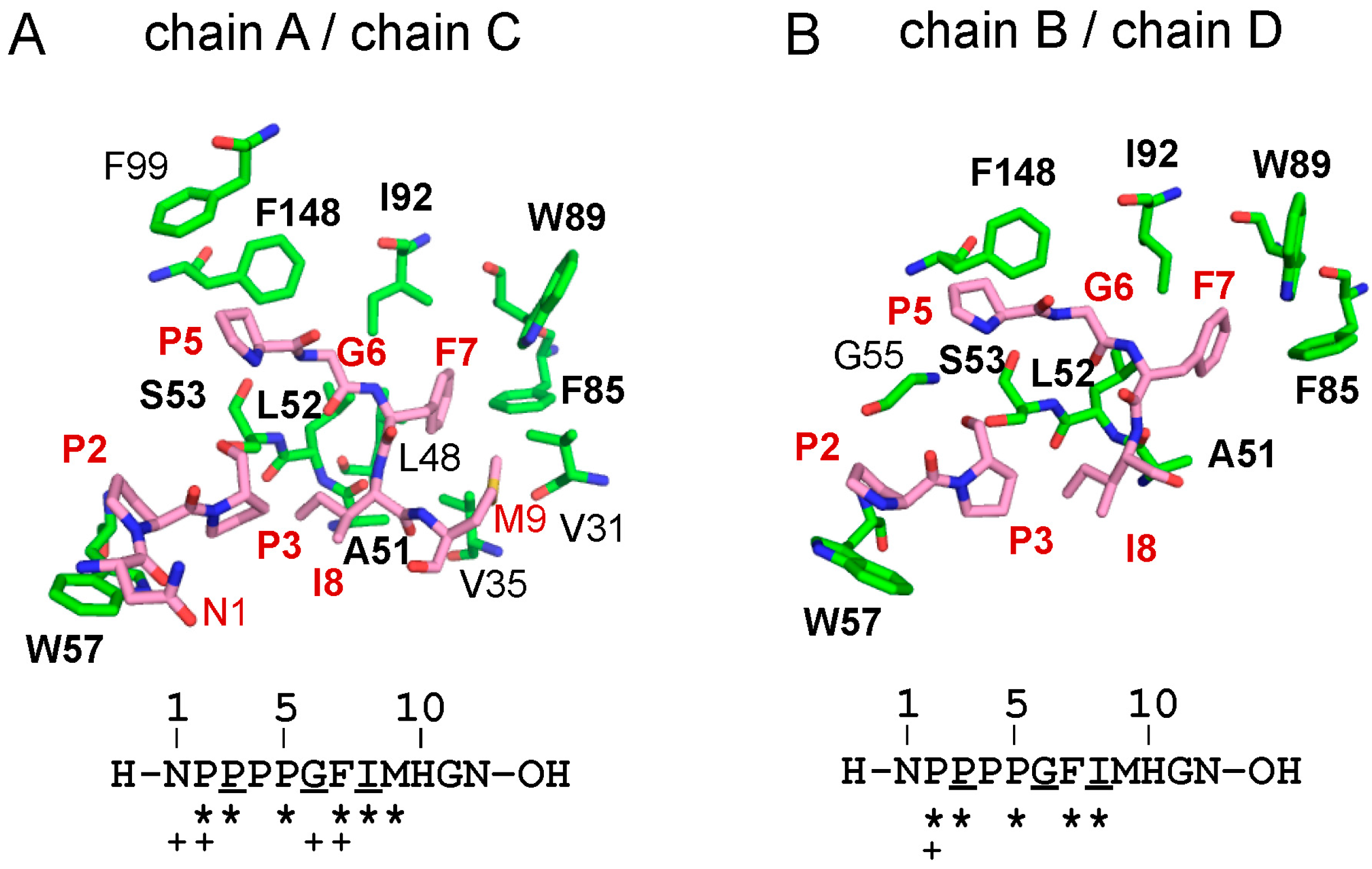
2.3. Structure of the Sec31A Peptide
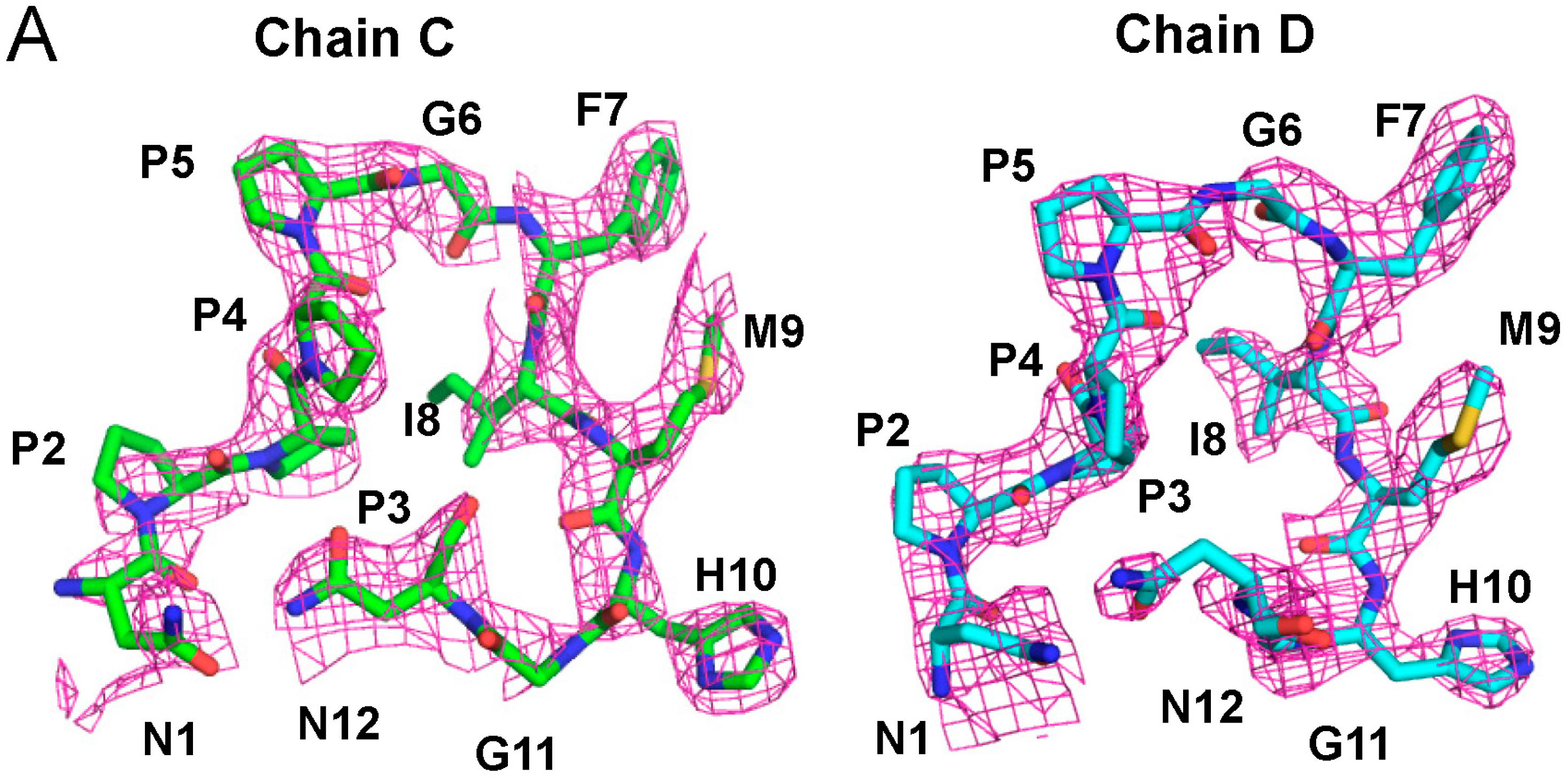
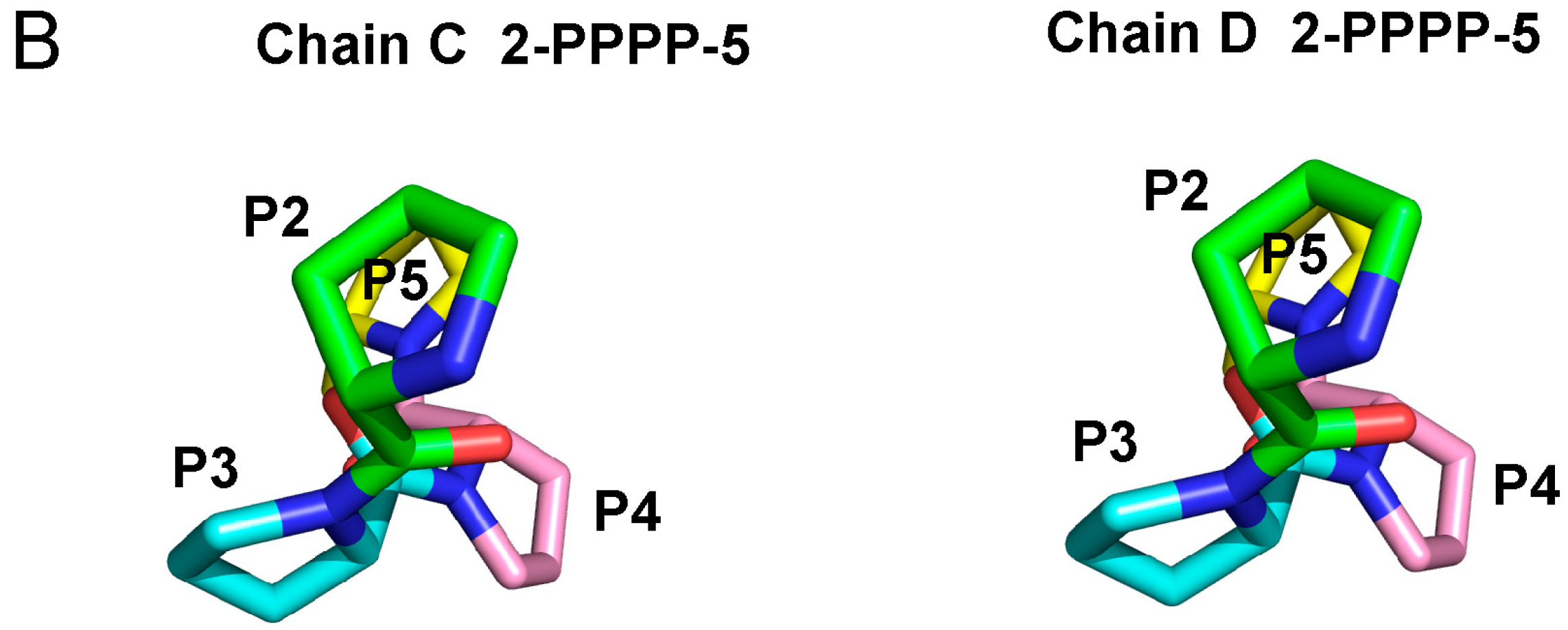
2.4. Binding of the Sec31A Peptide to Pocket 3
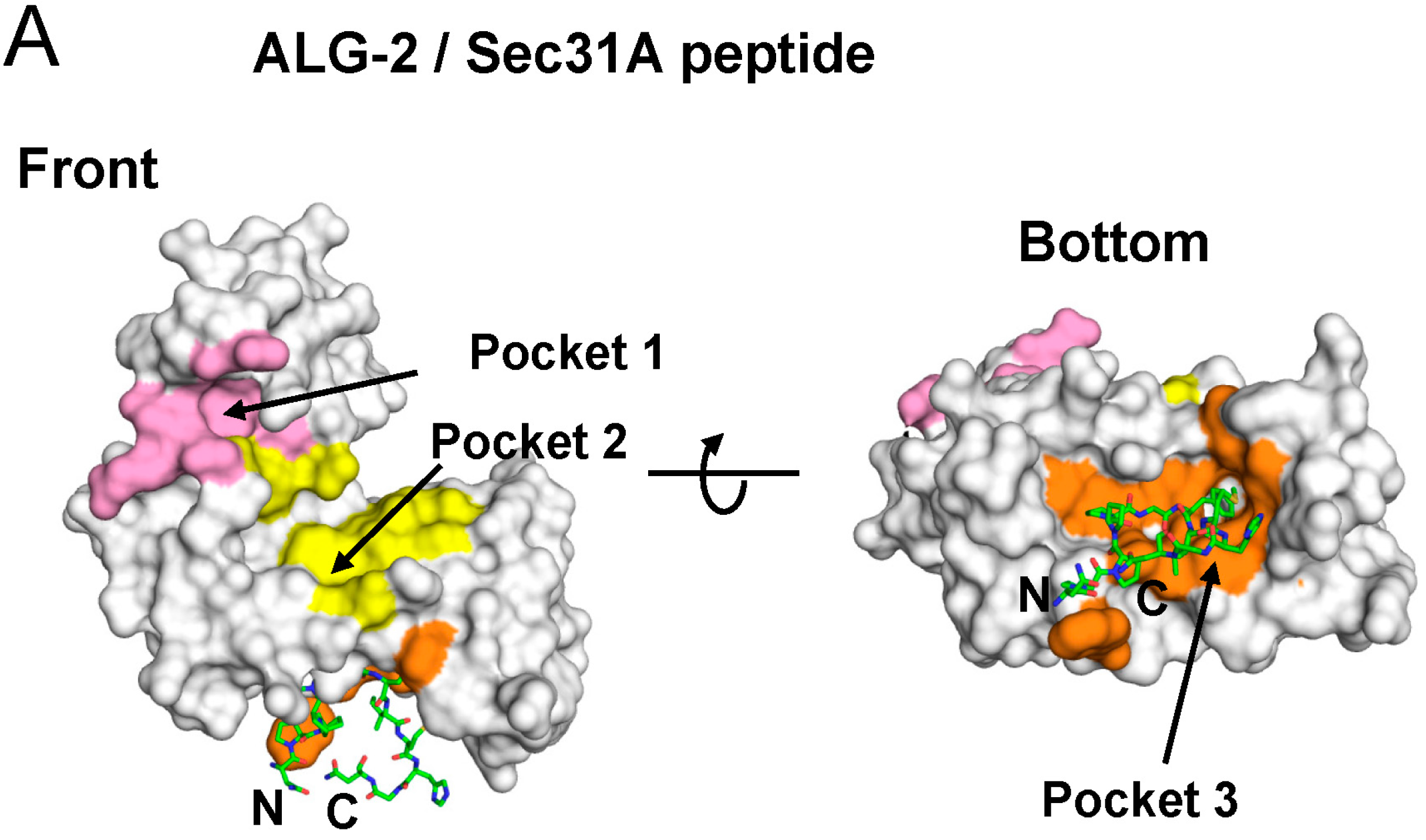
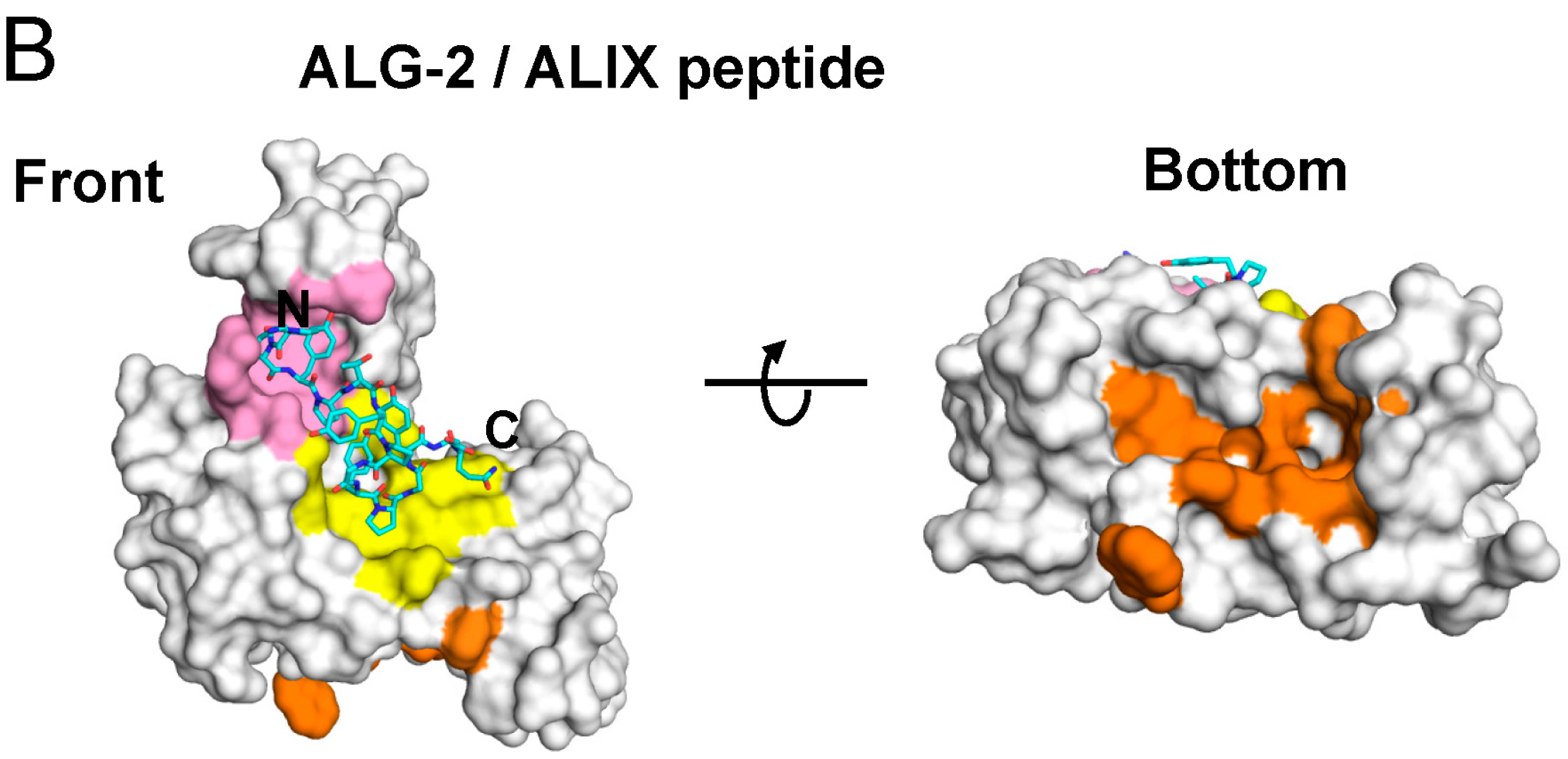
2.4.1. Analysis of the Ca2+-Dependent Interaction of Sec31A by GST-Pulldown
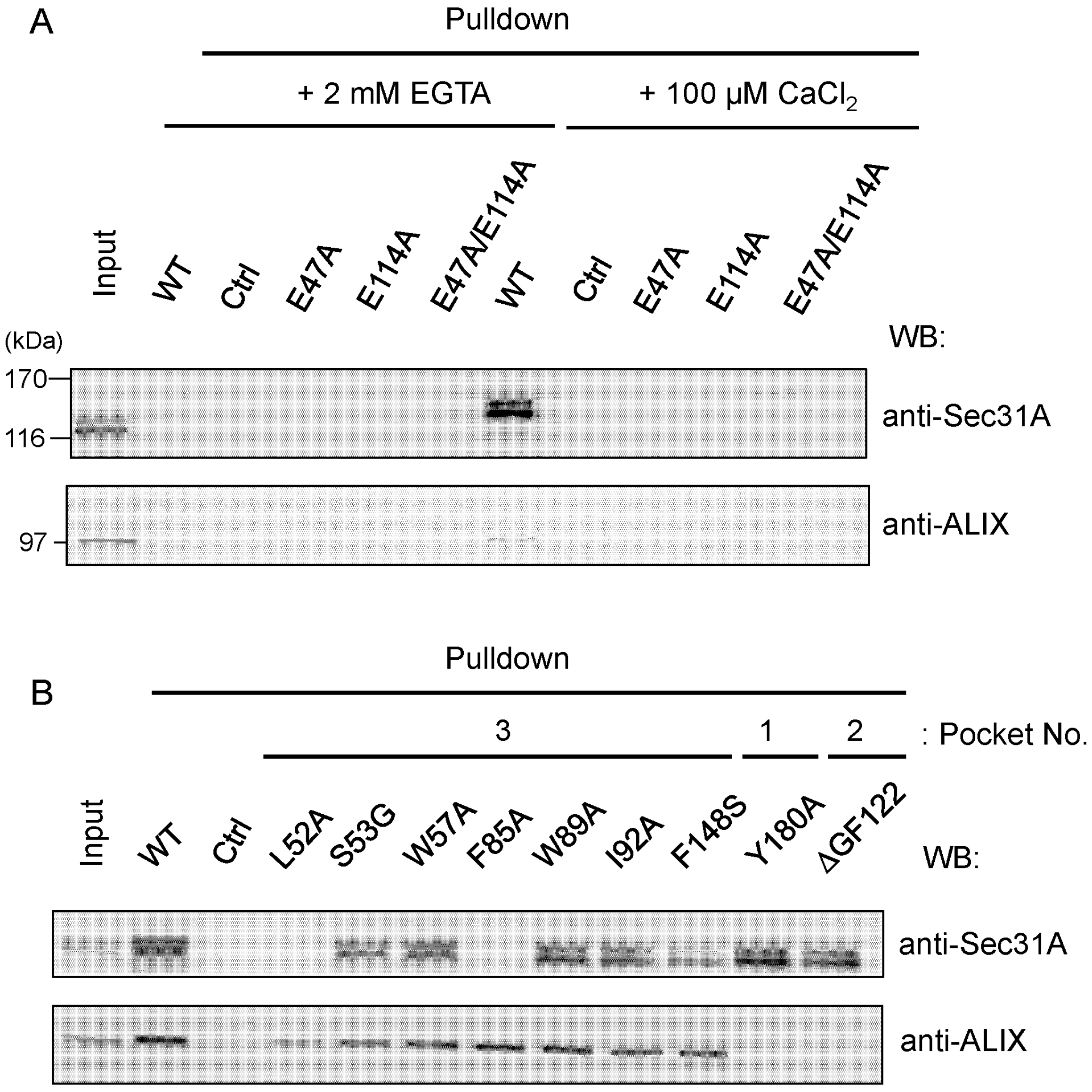
2.4.2. Identification of Critical Pocket 3 Residues for Interaction
2.5. Co-Immunoprecipitation Assays of Sec31A Mutants
2.6. Identification of Critical Residues in Sec31A for Binding to ALG-2

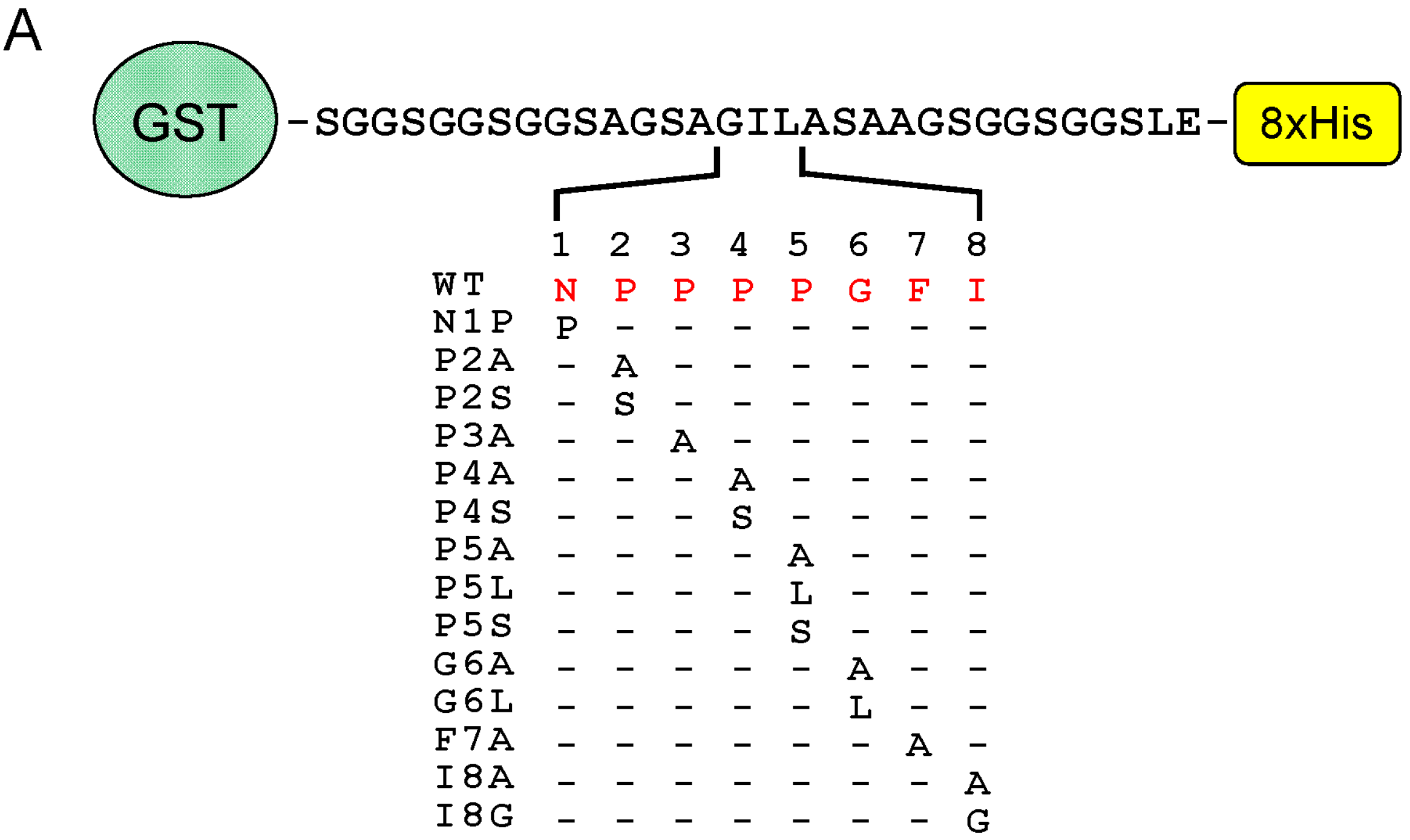

2.7. Comparison of ALG-2-Binding Motif Type 2 Sequences

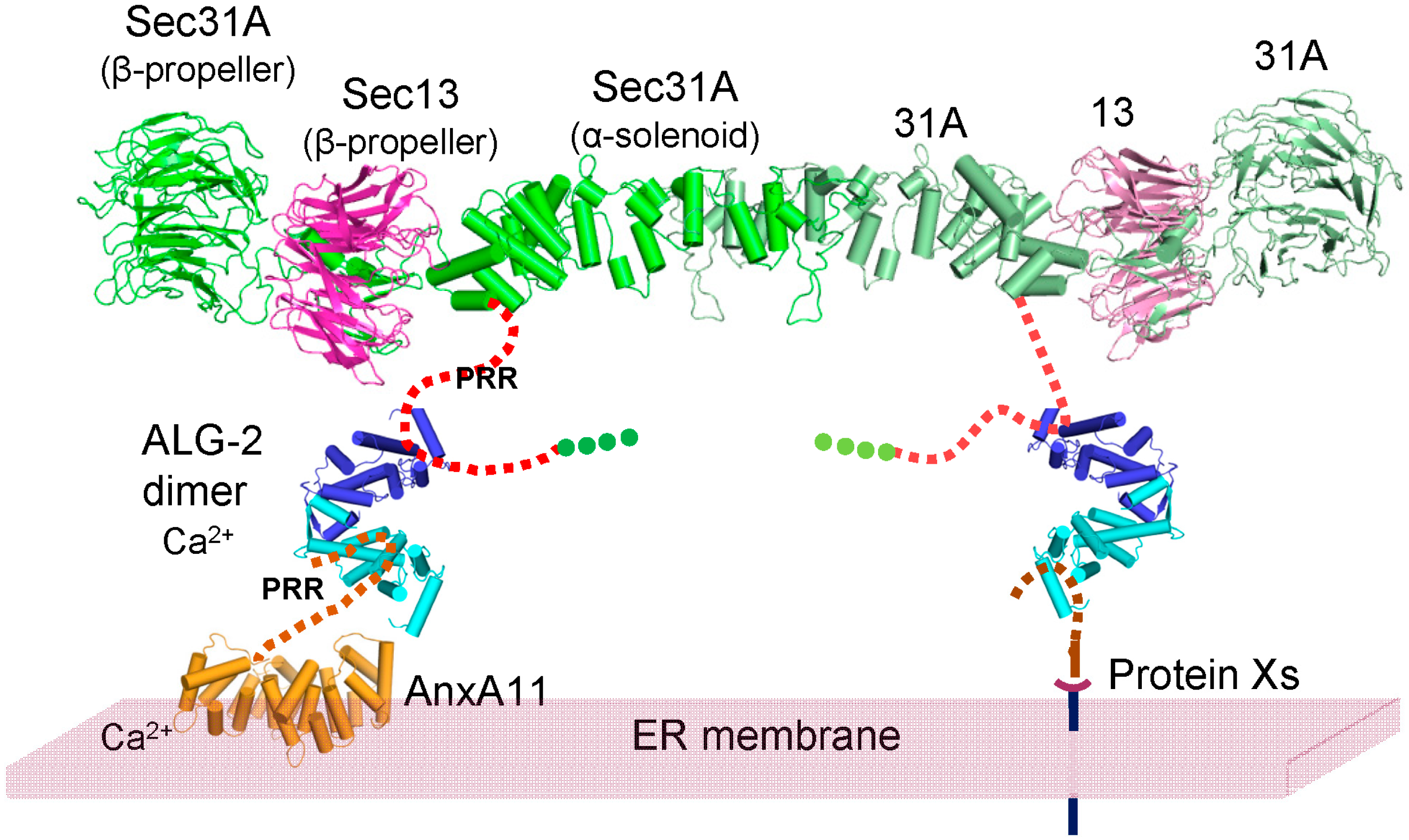
3. Discussion
4. Experimental Section
4.1. Expression Plasmids
4.2. Purification of Recombinant Proteins
4.3. Crystallization
4.4. Data Collection, Structure Determination and Refinement
4.5. Cell Culture and DNA Transfection
4.6. GST-ALG-2 Pulldown
4.7. Far-Western
4.8. Co-Immunoprecipitation Assays
4.9. Miscellaneous
Supplementary Materials
Acknowledgments
Author Contributions
Abbreviation
| aa | amino acids |
| Anx | annexin |
| bio-ALG-2 | biotin-labeled ALG-2 |
| COPII | coat protein complex II |
| ESCRT | endosomal sorting complex required for transport |
| FW | far-Western |
| GFP | green fluorescent protein |
| GST | glutathione-S-transferase |
| IP | immunoprecipitation |
| PEF | penta-EF-hand |
| PPII | polyproline helix type II |
| WB | Western blotting |
Conflicts of Interest
References
- Maki, M.; Kitaura, Y.; Satoh, H.; Ohkouchi, S.; Shibata, H. Structures, functions and molecular evolution of the penta-EF-hand Ca2+-binding proteins. Biochim. Biophys. Acta 2002, 1600, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Maki, M.; Maemoto, Y.; Osako, Y.; Shibata, H. Evolutionary and physical linkage between calpains and penta-EF-hand Ca2+-binding proteins. FEBS J. 2012, 279, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Vito, P.; Lacanà, E.; D’Adamio, L. Interfering with apoptosis: Ca2+-binding protein ALG-2 and Alzheimer’s disease gene ALG-3. Science 1996, 271, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.V.; Poksay, K.S.; Castro-Obregon, S.; Schilling, B.; Row, R.H.; del Rio, G.; Gibson, B.W.; Ellerby, H.M.; Bredesen, D.E. Molecular components of a cell death pathway activated by endoplasmic reticulum stress. J. Biol. Chem. 2004, 279, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Mahul-Mellier, A.L.; Strappazzon, F.; Petiot, A.; Chatellard-Causse, C.; Torch, S.; Blot, B.; Freeman, K.; Kuhn, L.; Garin, J.; Verna, J.M.; et al. Alix and ALG-2 are involved in tumor necrosis factor receptor 1-induced cell death. J. Biol. Chem. 2008, 283, 34954–34965. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Dashzeveg, N.; Lu, Z.G.; Taira, N.; Miki, Y.; Yoshida, K. Programmed cell death 6, a novel p53-responsive gene, targets to the nucleus in the apoptotic response to DNA damage. Cancer Sci. 2012, 103, 1788–1794. [Google Scholar] [CrossRef] [PubMed]
- La Cour, J.M.; Høj, B.R.; Mollerup, J.; Simon, R.; Sauter, G.; Berchtold, M.W. The apoptosis linked gene ALG-2 is dysregulated in tumors of various origin and contributes to cancer cell viability. Mol. Oncol. 2008, 1, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Arao, T.; Gotoda, T.; Taniguchi, H.; Oda, I.; Shirao, K.; Shimada, Y.; Hamaguchi, T.; Kato, K.; Hamano, T.; et al. Identification of prognostic biomarkers in gastric cancer using endoscopic biopsy samples. Cancer Sci. 2008, 99, 2193–2199. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Sytkowski, A.J. Apoptosis-linked gene-2 connects the Raf-1 and Ask1 signalings. Biochem. Biophys. Res. Commun. 2005, 333, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Rho, S.B.; Song, Y.J.; Lim, M.C.; Lee, S.H.; Kim, B.R.; Park, S.Y. Programmed cell death 6 (PDCD6) inhibits angiogenesis through PI3K/mTOR/p70S6K pathway by interacting of VEGFR-2. Cell Signal. 2012, 24, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, A.; Tani, K.; Yamamoto, A.; Kitamura, N.; Komada, M. The Ca2+-binding protein ALG-2 is recruited to endoplasmic reticulum exit sites by Sec31A and stabilizes the localization of Sec31A. Mol. Biol. Cell 2006, 17, 4876–4887. [Google Scholar] [CrossRef] [PubMed]
- Shibata, H.; Suzuki, H.; Yoshida, H.; Maki, M. ALG-2 directly binds SEC31A and localizes at endoplasmic reticulum exit sites in a Ca2+-dependent manner. Biochem. Biophys. Res. Commun. 2007, 353, 756–763. [Google Scholar] [CrossRef] [PubMed]
- La Cour, J.M.; Mollerup, J.; Berchtold, M.W. ALG-2 oscillates in subcellular localization, unitemporally with calcium oscillations. Biochem. Biophys. Res. Commun. 2007, 353, 1063–1067. [Google Scholar] [CrossRef] [PubMed]
- Bentley, M.; Nycz, D.C.; Joglekar, A.; Fertschai, I.; Malli, R.; Graier, W.F.; Hay, J.C. Vesicular calcium regulates coat retention, fusogenicity, and size of pre-Golgi intermediates. Mol. Biol. Cell 2010, 21, 1033–1046. [Google Scholar] [CrossRef] [PubMed]
- Krebs, J. The influence of calcium signaling on the regulation of alternative splicing. Biochim. Biophys. Acta 2009, 1793, 979–984. [Google Scholar] [CrossRef] [PubMed]
- Sasaki-Osugi, K.; Imoto, C.; Takahara, T.; Shibata, H.; Maki, M. Nuclear ALG-2 protein interacts with Ca2+ homeostasis endoplasmic reticulum protein (CHERP) Ca2+-dependently and participates in regulation of alternative splicing of inositol trisphosphate receptor type 1 (IP3R1) pre-mRNA. J. Biol. Chem. 2013, 288, 33361–33375. [Google Scholar] [CrossRef] [PubMed]
- Maki, M.; Suzuki, H.; Shibata, H. Structure and function of ALG-2, a penta-EF-hand calcium-dependent adaptor protein. Sci. China Life Sci. 2011, 54, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Osugi, K.; Suzuki, H.; Nomura, T.; Ariumi, Y.; Shibata, H.; Maki, M. Identification of the P-body component PATL1 as a novel ALG-2-interacting protein by in silico and far-Western screening of proline-rich proteins. J. Biochem. 2012, 151, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, G.; Pahuja, K.B.; Studer, S.; Shim, S.; Schekman, R. COPII and the regulation of protein sorting in mammals. Nat. Cell Biol. 2012, 14, 20–28. [Google Scholar] [CrossRef]
- Venditti, R.; Wilson, C.; de Matteis, M.A. Exiting the ER: What we know and what we don’t. Trends Cell Biol. 2014, 24, 9–18. [Google Scholar] [CrossRef] [PubMed]
- La Cour, J.M.; Schindler, A.J.; Berchtold, M.W.; Schekman, R. ALG-2 attenuates COPII budding in vitro and stabilizes the Sec23/Sec31A complex. PLoS One 2013, 8, e75309. [Google Scholar] [CrossRef] [PubMed]
- Helm, J.R.; Bentley, M.; Thorsen, K.D.; Wang, T.; Foltz, L.; Oorschot, V.; Klumperman, J.; Hay, J.C. Apoptosis-linked gene-2 (ALG-2)/Sec31 interactions regulate endoplasmic reticulum (ER)-to-Golgi transport: A potential effector pathway for luminal calcium. J. Biol. Chem. 2014, 289, 23609–23628. [Google Scholar] [CrossRef] [PubMed]
- Shibata, H.; Kanadome, T.; Sugiura, H.; Yokoyama, T.; Yamamuro, M.; Moss, S.E.; Maki, M. A new role for annexin A11 in the early secretory pathway via stabilizing Sec31A protein at the endoplasmic reticulum exit sites (ERES). J. Biol. Chem. 2015, 290, 4981–4993. [Google Scholar]
- Missotten, M.; Nichols, A.; Rieger, K.; Sadoul, R. Alix, a novel mouse protein undergoing calcium-dependent interaction with the apoptosis-linked-gene 2 (ALG-2) protein. Cell Death Differ. 1999, 6, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Vito, P.; Pellegrini, L.; Guiet, C.; D’Adamio, L. Cloning of AIP1, a novel protein that associates with the apoptosis-linked gene ALG-2 in a Ca2+-dependent reaction. J. Biol. Chem. 1999, 274, 1533–1540. [Google Scholar] [CrossRef] [PubMed]
- Shibata, H.; Yamada, K.; Mizuno, T.; Yorikawa, C.; Takahashi, H.; Satoh, H.; Kitaura, Y.; Maki, M. The penta-EF-hand protein ALG-2 interacts with a region containing PxY repeats in Alix/AIP1, which is required for the subcellular punctate distribution of the amino-terminal truncation form of Alix/AIP1. J. Biochem. 2004, 135, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Kawasaki, M.; Inuzuka, T.; Okumura, M.; Kakiuchi, T.; Shibata, H.; Wakatsuki, S.; Maki, M. Structural basis for Ca2+-dependent formation of ALG-2/Alix peptide complex: Ca2+/EF3-driven arginine switch mechanism. Structure 2008, 16, 1562–1573. [Google Scholar] [CrossRef] [PubMed]
- Shibata, H.; Suzuki, H.; Kakiuchi, T.; Inuzuka, T.; Yoshida, H.; Mizuno, T.; Maki, M. Identification of Alix-type and non-Alix-type ALG-2-binding sites in human phospholipid scramblase 3: Differential binding to an alternatively spliced isoform and amino acid-substituted mutants. J. Biol. Chem. 2008, 283, 9623–9632. [Google Scholar] [CrossRef] [PubMed]
- Shibata, H.; Inuzuka, T.; Yoshida, H.; Sugiura, H.; Wada, I.; Maki, M. The ALG-2 binding site in Sec31A influences the retention kinetics of Sec31A at the endoplasmic reticulum exit sites as revealed by live-cell time-lapse imaging. Biosci. Biotechnol. Biochem. 2010, 74, 1819–1826. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Suzuki, H.; Inuzuka, T.; Shibata, H.; Maki, M. Prediction of a new ligand-binding site for type 2 motif based on the crystal structure of ALG-2 by dry and wet approaches. Int. J. Mol. Sci. 2012, 13, 7532–7549. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Tarabykina, S.; Hansen, C.; Berchtold, M.; Cygler, M. Structure of apoptosis-linked protein ALG-2: Insights into Ca2+-induced changes in penta-EF-hand proteins. Structure 2001, 9, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Inuzuka, T.; Suzuki, H.; Kawasaki, M.; Shibata, H.; Wakatsuki, S.; Maki, M. Molecular basis for defect in Alix-binding by alternatively spliced isoform of ALG-2 (ALG-2ΔGF122) and structural roles of F122 in target recognition. BMC Struct. Biol. 2010, 10, 25. [Google Scholar] [PubMed]
- Feng, S.; Chen, J.K.; Yu, H.; Simon, J.A.; Schreiber, S.L. Two binding orientations for peptides to the Src SH3 domain: Development of a general model for SH3-ligand interactions. Science 1994, 266, 1241–1247. [Google Scholar] [CrossRef] [PubMed]
- Alexandropoulos, K.; Cheng, G.; Baltimore, D. Proline-rich sequences that bind to Src homology 3 domains with individual specificities. Proc. Natl. Acad. Sci. USA 1995, 92, 3110–3114. [Google Scholar] [CrossRef] [PubMed]
- Sudol, M.; Hunter, T. New wrinkles for an old domain. Cell 2000, 103, 1001–1004. [Google Scholar] [CrossRef] [PubMed]
- Okumura, M.; Ichioka, F.; Kobayashi, R.; Suzuki, H.; Yoshida, H.; Shibata, H.; Maki, M. Penta-EF-hand protein ALG-2 functions as a Ca2+-dependent adaptor that bridges ALIX and TSG101. Biochem. Biophys. Res. Commun. 2009, 386, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Kawasaki, M.; Kakiuchi, T.; Shibata, H.; Wakatsuki, S.; Maki, M. Crystallization and X-ray diffraction analysis of N-terminally truncated human ALG-2. Acta Crystallogr. 2008, 64, 974–977. [Google Scholar]
- Maki, M.; Narayana, S.V.; Hitomi, K. A growing family of the Ca2+-binding proteins with five EF-hand motifs. Biochem. J. 1997, 328, 718–720. [Google Scholar] [PubMed]
- Katoh, K.; Suzuki, H.; Terasawa, Y.; Mizuno, T.; Yasuda, J.; Shibata, H.; Maki, M. The penta-EF-hand protein ALG-2 interacts directly with the ESCRT-I component TSG101, and Ca2+-dependently co-localizes to aberrant endosomes with dominant-negative AAA ATPase SKD1/Vps4B. Biochem. J. 2005, 391, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Okumura, M.; Katsuyama, A.M.; Shibata, H.; Maki, M. VPS37 isoforms differentially modulate the ternary complex formation of ALIX, ALG-2, and ESCRT-I. Biosci. Biotechnol. Biochem. 2013, 77, 1715–1721. [Google Scholar] [CrossRef] [PubMed]
- Stagg, S.M.; Gurkan, C.; Fowler, D.M.; LaPointe, P.; Foss, T.R.; Potter, C.S.; Carragher, B.; Balch, W.E. Structure of the Sec13/31 COPII coat cage. Nature 2006, 439, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Noble, A.J.; Zhang, Q.; O’Donnell, J.; Hariri, H.; Bhattacharya, N.; Marshall, A.G.; Stagg, S.M. A pseudoatomic model of the COPII cage obtained from cryo-electron microscopy and mass spectrometry. Nat. Struct. Mol. Biol. 2013, 20, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, G.; Prinz, S.; Daum, S.; Meister, A.; Schekman, R.; Bacia, K.; Briggs, J.A. The structure of the COPII transport-vesicle coat assembled on membranes. Elife 2013, 2, e00951. [Google Scholar] [CrossRef] [PubMed]
- Fath, S.; Mancias, J.D.; Bi, X.; Goldberg, J. Structure and organization of coat proteins in the COPII cage. Cell 2007, 129, 1325–1336. [Google Scholar] [CrossRef] [PubMed]
- Vernarecci, S.; Colotti, G.; Ornaghi, P.; Schiebel, E.; Chiancone, E.; Filetici, P. The yeast penta-EFprotein Pef1p is involved in cation-dependent budding and cell polarization. Mol. Microbiol. 2007, 65, 1122–1138. [Google Scholar] [CrossRef] [PubMed]
- Yoshibori, M.; Yorimitsu, T.; Sato, K. Involvement of the penta-EF-hand protein Pef1p in the Ca2+-dependent regulation of COPII subunit assembly in Saccharomyces cerevisiae. PLoS One 2012, 7, e40765. [Google Scholar] [CrossRef] [PubMed]
- Lollike, K.; Johnsen, A.H.; Durussel, I.; Borregaard, N.; Cox, J.A. Biochemical characterization of the penta-EF-hand protein grancalcin and identification of L-plastin as a binding partner. J. Biol. Chem. 2001, 276, 17762–17769. [Google Scholar] [CrossRef] [PubMed]
- Braun, E.L.; Kang, S.; Nelson, M.A.; Natvig, D.O. Identification of the first fungal annexin: Analysis of annexin gene duplications and implications for eukaryotic evolution. J. Mol. Evol. 1998, 47, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Zacharias, D.A.; Violin, J.D.; Newton, A.C.; Tsien, R.Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 2002, 296, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Hiraki, M.; Kato, R.; Nagai, M.; Satoh, T.; Hirano, S.; Ihara, K.; Kudo, N.; Nagae, M.; Kobayashi, M.; Inoue, M.; et al. Development of an automated large-scale protein-crystallization and monitoring system for high-throughput protein-structure analyses. Acta Crystallogr. 2006, 62, 1058–1065. [Google Scholar]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]
- Vagin, A.; Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. 2010, 66, 22–25. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997, 53, 240–255. [Google Scholar]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004, 60, 2126–2132. [Google Scholar]
- Osugi, K.; Shibata, H.; Maki, M. Biochemical and immunological detection of physical interactions between penta-EF-hand protein ALG-2 and its binding partners. Methods Mol. Biol. 2013, 963, 187–200. [Google Scholar] [PubMed]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- SWISS-MODEL. Available online: http://swissmodel.expasy.org/ (accessed on 27 December 2014).
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Clustal Omega. Available online: http://www.ebi.ac.uk/Tools/msa/clustalo/ (accessed on 29 December 2014).
- Color h. Available online: http://pymolwiki.org/index.php/Color_h (accessed on 22 January 2015).
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, T.; Kojima, K.; Zhang, W.; Sasaki, K.; Ito, M.; Suzuki, H.; Kawasaki, M.; Wakatsuki, S.; Takahara, T.; Shibata, H.; et al. Structural Analysis of the Complex between Penta-EF-Hand ALG-2 Protein and Sec31A Peptide Reveals a Novel Target Recognition Mechanism of ALG-2. Int. J. Mol. Sci. 2015, 16, 3677-3699. https://doi.org/10.3390/ijms16023677
Takahashi T, Kojima K, Zhang W, Sasaki K, Ito M, Suzuki H, Kawasaki M, Wakatsuki S, Takahara T, Shibata H, et al. Structural Analysis of the Complex between Penta-EF-Hand ALG-2 Protein and Sec31A Peptide Reveals a Novel Target Recognition Mechanism of ALG-2. International Journal of Molecular Sciences. 2015; 16(2):3677-3699. https://doi.org/10.3390/ijms16023677
Chicago/Turabian StyleTakahashi, Takeshi, Kyosuke Kojima, Wei Zhang, Kanae Sasaki, Masaru Ito, Hironori Suzuki, Masato Kawasaki, Soichi Wakatsuki, Terunao Takahara, Hideki Shibata, and et al. 2015. "Structural Analysis of the Complex between Penta-EF-Hand ALG-2 Protein and Sec31A Peptide Reveals a Novel Target Recognition Mechanism of ALG-2" International Journal of Molecular Sciences 16, no. 2: 3677-3699. https://doi.org/10.3390/ijms16023677
APA StyleTakahashi, T., Kojima, K., Zhang, W., Sasaki, K., Ito, M., Suzuki, H., Kawasaki, M., Wakatsuki, S., Takahara, T., Shibata, H., & Maki, M. (2015). Structural Analysis of the Complex between Penta-EF-Hand ALG-2 Protein and Sec31A Peptide Reveals a Novel Target Recognition Mechanism of ALG-2. International Journal of Molecular Sciences, 16(2), 3677-3699. https://doi.org/10.3390/ijms16023677