The Mechanisms of Virulence Regulation by Small Noncoding RNAs in Low GC Gram-Positive Pathogens
Abstract
:1. Introduction
| Bacteria | Small RNA | Target mRNA | Protein Encoded in Target mRNA | References |
|---|---|---|---|---|
| S. pyogenes | PelRNA | speB | Cysteine protease | [1] |
| emm | M protein | |||
| sic | Streptococcal inhibitor of complement | |||
| nga | NAD-glycohydrolase | |||
| FasX | ska | Streptokinase | [9,10] | |
| sagS | Streptolysin S | |||
| fbp54 | Fibronectin binding protein | |||
| mrp | M-related protein | |||
| RivX | mga | Virulence regulator | [11] | |
| S. pneumoniae | F20 | Not known | – | [12] |
| F32 | – | F32 encodes a tmRNA | [12] | |
| S. aureus | RNAIII | hla | α-hemolysin | [1,2,13] |
| rot | Repressor of toxins | [14,15,16] | ||
| sa1000 | Fibrinogen-binding protein | [1,15] | ||
| spa | Protein A | [17] | ||
| coa | Coagulase | [18] | ||
| lytM | Peptidoglycan hydrolase | [19] | ||
| map | MHCII analogous protein | [20,21] | ||
| SprD | sbi | Immunoglobulin-binding protein | [22] | |
| Psm-mec | agrA | Virulence regulator | [23] | |
| ArtR | sarT | Transcriptional regulator of hla | [24] | |
| L. monocytogenes | SreA | prfA | Virulence master regulator | [25] |
| agrD | Precusor of autoinducer | |||
| SreB | prfA | Virulence master regulator | [25] | |
| LhrA | chiA | Chitinase | [26] | |
| LhrC | lapB | Virulence adhesin | [6,27] | |
| C. perfringens | VR-RNA | colA | Collagenase | [28,29,30,31] |
| pfoA | Pore-forming toxin Perfringolysin A | |||
| vrr | VirRS-regulated sRNA | |||
| virT | The VirT sRNA | |||
| virU | The VirU sRNA | |||
| ccp | Cysteine protease α-clostripain | |||
| VirX | pfoA | Pore-forming toxin Perfringolysin A | [31] | |
| plc | Alpha-toxin phospholipase C | |||
| colA | Collagenase | |||
| VirU | vrr | VirRS-regulated sRNA | [31] | |
| pfoA | Pore-forming toxin Perfringolysin A | |||
| virT | The VirT sRNA | |||
| VirT | pfoA | Pore-forming toxin Perfringolysin A | [31] | |
| colA | Collagenase | |||
| E. faecalis | Ef0408-0409 | Fst | Peptide toxin | [32] |
1.1. Streptococcus pyogenes
1.2. Streptococcus pneumoniae
1.3. Staphylococcus aureus
1.4. Listeria monocytogenes
1.5. Clostridium perfringens
1.6. Enterococcus faecalis
2. Common Features Shared by sRNAs in Gram-Positive Pathogens
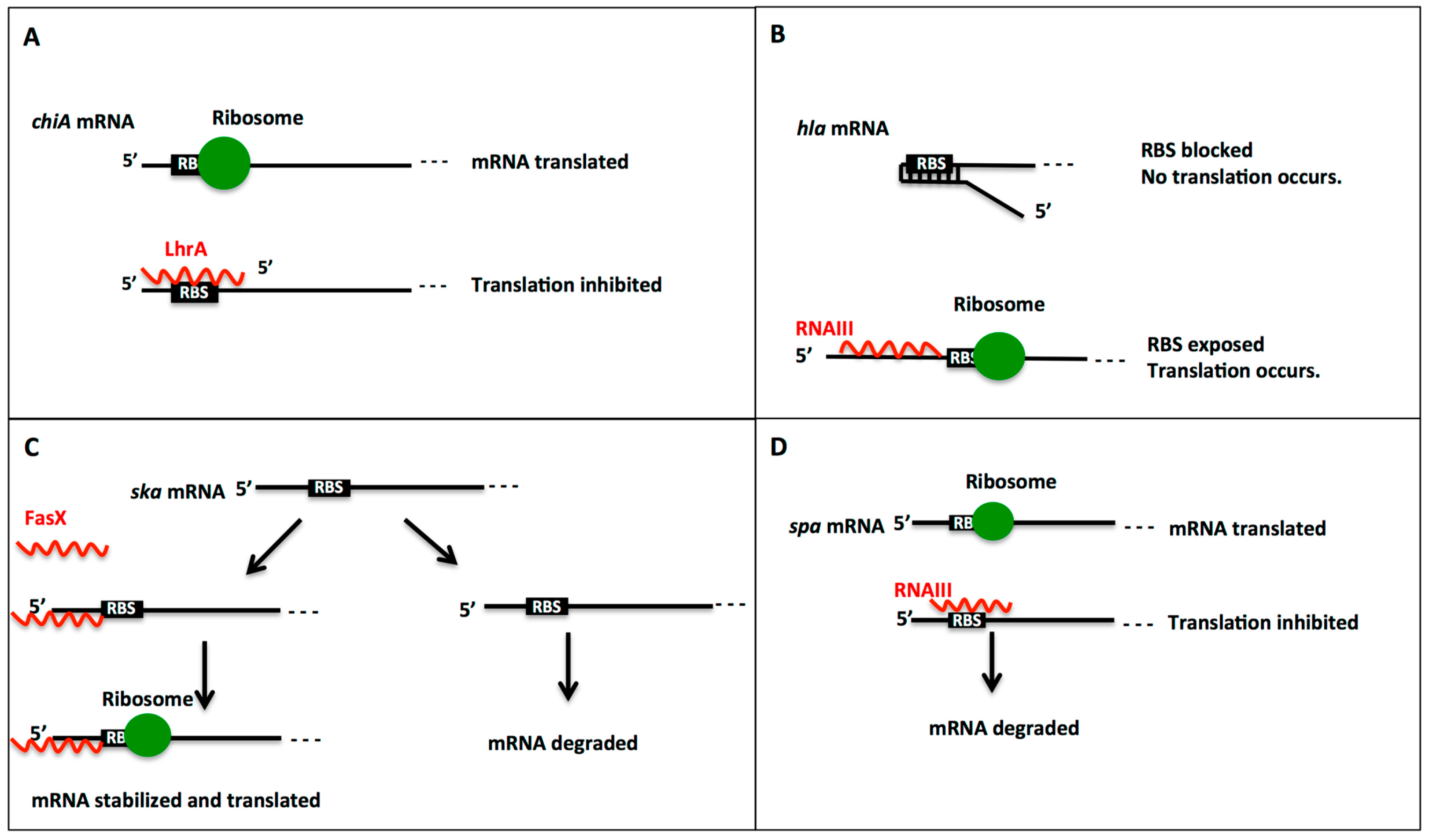
2.1. Regulation Mechanisms by Small RNAs: Activation or Repression of Gene Expression
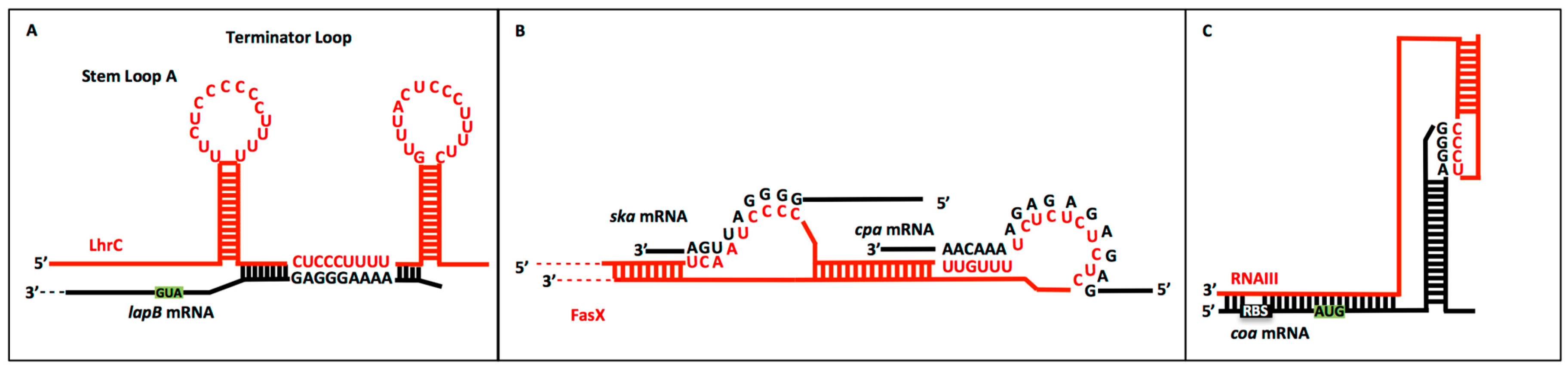
2.2. CU Repeats in sRNA:Target mRNA Interactions
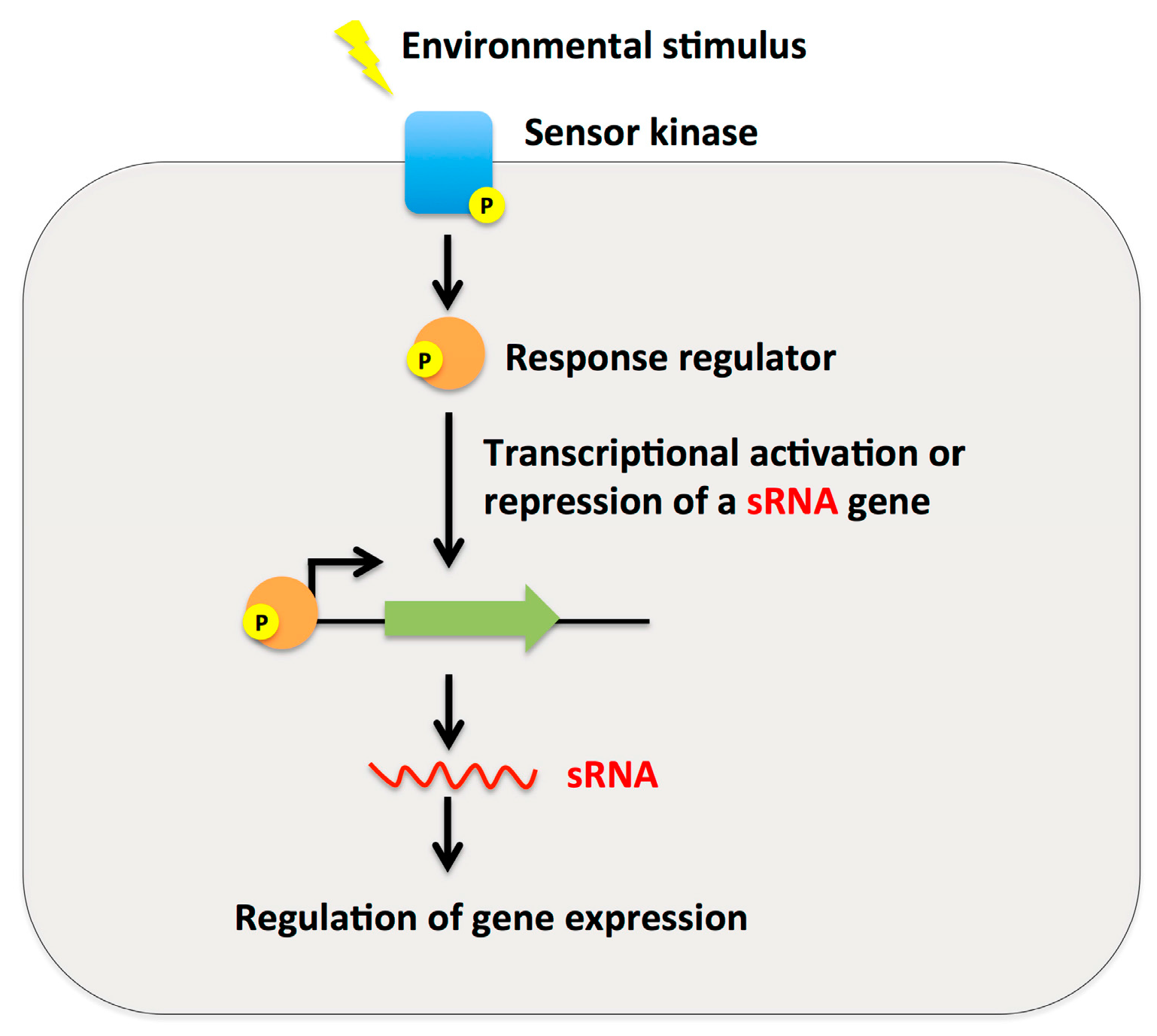
2.3. Small RNAs as Effector Molecules of Two-Component Systems
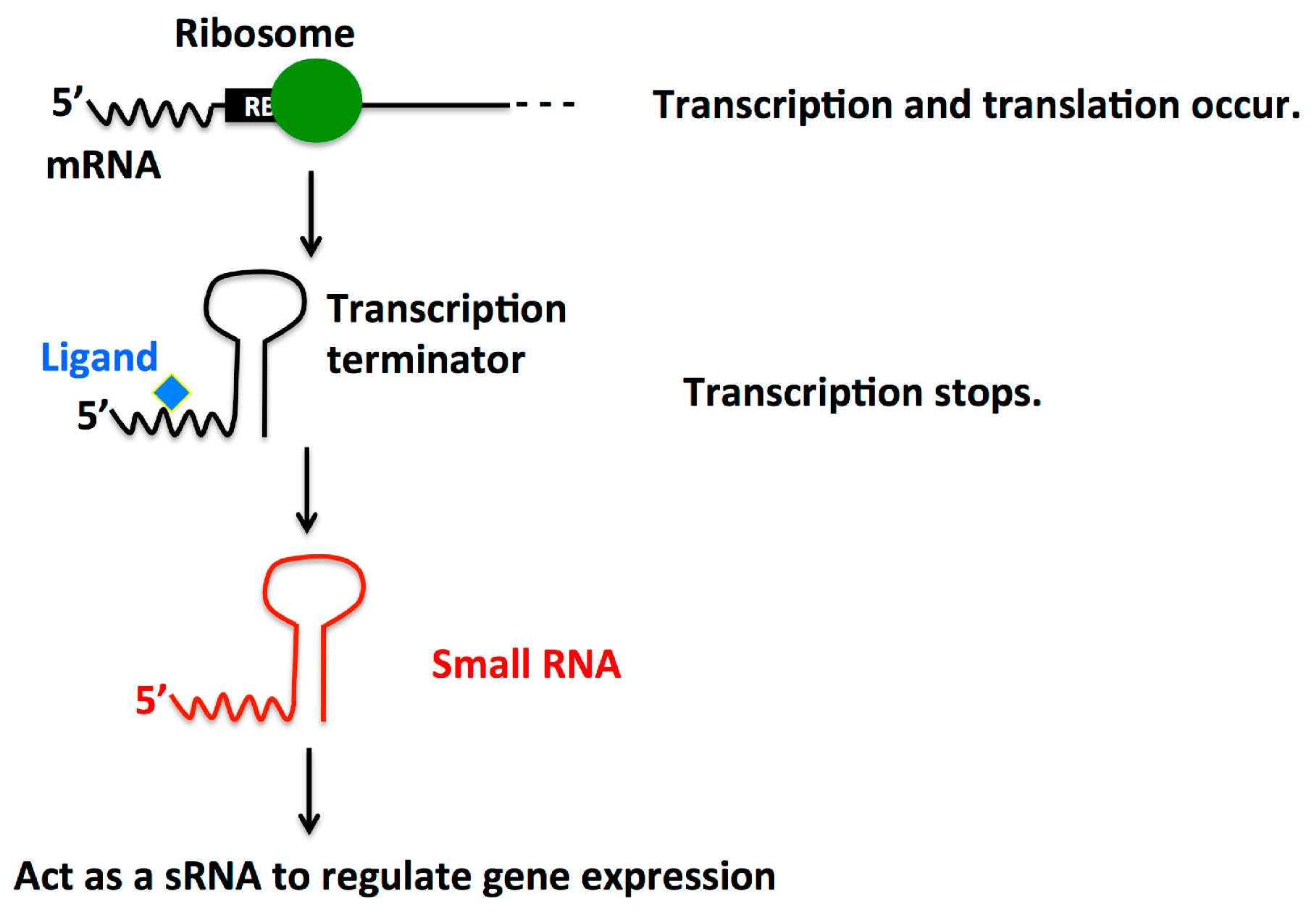
2.4. Dual Function Riboswitch/sRNA
2.5. Small RNAs in Quorum Sensing System

2.6. Small RNAs in Toxin/Antitoxin System
2.7. The Involvement of Hfq in sRNA Activity
3. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Brantl, S.; Bruckner, R. Small regulatory RNAs from low-GC gram-positive bacteria. RNA Biol. 2014, 11, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.W.; Cao, T.N.; Pflughoeft, K.J.; Sumby, P. RNA-mediated regulation in gram-positive pathogens: An overview punctuated with examples from the Group A Streptococcus. Mol. Microbiol. 2014, 94, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.H.; Kim, J.H. Cis-encoded non-coding antisense RNAs in streptococci and other low GC gram (+) bacterial pathogens. Front. Genet. 2015, 6, 110. [Google Scholar] [CrossRef] [PubMed]
- Pichon, C.; Felden, B. Small RNA gene identification and mRNA target predictions in bacteria. Bioinformatics 2008, 24, 2807–2813. [Google Scholar] [CrossRef] [PubMed]
- Tesorero, R.A.; Yu, N.; Wright, J.O.; Svencionis, J.P.; Cheng, Q.; Kim, J.H.; Cho, K.H. Novel regulatory small RNAs in Streptococcus pyogenes. PLoS ONE 2013, 8, e64021. [Google Scholar] [CrossRef] [PubMed]
- Sievers, S.; Sternkopf Lillebaek, E.M.; Jacobsen, K.; Lund, A.; Mollerup, M.S.; Nielsen, P.K.; Kallipolitis, B.H. A multicopy sRNA of Listeria monocytogenes regulates expression of the virulence adhesin LapB. Nucleic Acids Res. 2014, 42, 9383–9398. [Google Scholar] [CrossRef] [PubMed]
- Johansson, J.; Mandin, P.; Renzoni, A.; Chiaruttini, C.; Springer, M.; Cossart, P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 2002, 110, 551–561. [Google Scholar] [CrossRef]
- Behrens, S.; Widder, S.; Mannala, G.K.; Qing, X.; Madhugiri, R.; Kefer, N.; Abu Mraheil, M.; Rattei, T.; Hain, T. Ultra deep sequencing of Listeria monocytogenes sRNA transcriptome revealed new antisense RNAs. PLoS ONE 2014, 9, e83979. [Google Scholar] [CrossRef] [PubMed]
- Kreikemeyer, B.; Boyle, M.D.; Buttaro, B.A.; Heinemann, M.; Podbielski, A. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol. Microbiol. 2001, 39, 392–406. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Pena, E.; Trevino, J.; Liu, Z.; Perez, N.; Sumby, P. The Group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol. Microbiol. 2010, 78, 1332–1347. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.A.; Scott, J.R. RivR and the small RNA RivX: The missing links between the CovR regulatory cascade and the Mga regulon. Mol. Microbiol. 2007, 66, 1506–1522. [Google Scholar] [CrossRef] [PubMed]
- Mann, B.; van Opijnen, T.; Wang, J.; Obert, C.; Wang, Y.D.; Carter, R.; McGoldrick, D.J.; Ridout, G.; Camilli, A.; Tuomanen, E.I.; et al. Control of virulence by small RNAs in Streptococcus pneumoniae. PLoS Pathog. 2012, 8, e1002788. [Google Scholar] [CrossRef] [PubMed]
- Morfeldt, E.; Taylor, D.; von Gabain, A.; Arvidson, S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995, 14, 4569–4577. [Google Scholar] [PubMed]
- Novick, R.P.; Jiang, D. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 2003, 149, 2709–2717. [Google Scholar] [CrossRef] [PubMed]
- Boisset, S.; Geissmann, T.; Huntzinger, E.; Fechter, P.; Bendridi, N.; Possedko, M.; Chevalier, C.; Helfer, A.C.; Benito, Y.; Jacquier, A.; et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007, 21, 1353–1366. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Boisset, S.; Saveanu, C.; Benito, Y.; Geissmann, T.; Namane, A.; Lina, G.; Etienne, J.; Ehresmann, B.; Ehresmann, C.; et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005, 24, 824–835. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, C.; Boisset, S.; Romilly, C.; Masquida, B.; Fechter, P.; Geissmann, T.; Vandenesch, F.; Romby, P. Staphylococcus aureus RNAIII binds to two distant regions of coa mRNA to arrest translation and promote mRNA degradation. PLoS Pathog. 2010, 6, e1000809. [Google Scholar] [CrossRef] [PubMed]
- Chunhua, M.; Yu, L.; Yaping, G.; Jie, D.; Qiang, L.; Xiaorong, T.; Guang, Y. The expression of lytM is down-regulated by RNAIII in Staphylococcus aureus. J. Basic Microbiol. 2012, 52, 636–641. [Google Scholar] [CrossRef] [PubMed]
- Harraghy, N.; Hussain, M.; Haggar, A.; Chavakis, T.; Sinha, B.; Herrmann, M.; Flock, J.I. The adhesive and immunomodulating properties of the multifunctional Staphylococcus aureus protein Eap. Microbiology 2003, 149, 2701–2707. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Mu, C.; Ying, X.; Li, W.; Wu, N.; Dong, J.; Gao, Y.; Shao, N.; Fan, M.; Yang, G. RNAIII activates map expression by forming an RNA-RNA complex in Staphylococcus aureus. FEBS Lett. 2011, 585, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Zhang, X.; Sun, H.; Sun, B. ArtR, a novel sRNA of Staphylococcus aureus, regulates α-toxin expression by targeting the 5′-UTR of sarT mRNA. Med. Microbiol. Immunol. 2014, 203, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pichon, C.; Felden, B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. USA 2005, 102, 14249–14254. [Google Scholar] [CrossRef] [PubMed]
- Masse, E.; Majdalani, N.; Gottesman, S. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 2003, 6, 120–124. [Google Scholar] [CrossRef]
- Kaito, C.; Saito, Y.; Ikuo, M.; Omae, Y.; Mao, H.; Nagano, G.; Fujiyuki, T.; Numata, S.; Han, X.; Obata, K.; et al. Mobile genetic element SCCmec-encoded psm-mec RNA suppresses translation of agrA and attenuates MRSA virulence. PLoS Pathog. 2013, 9, e1003269. [Google Scholar] [CrossRef] [PubMed]
- Loh, E.; Dussurget, O.; Gripenland, J.; Vaitkevicius, K.; Tiensuu, T.; Mandin, P.; Repoila, F.; Buchrieser, C.; Cossart, P.; Johansson, J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 2009, 139, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.S.; Larsen, M.H.; Lillebaek, E.M.; Bergholz, T.M.; Christiansen, M.H.; Boor, K.J.; Wiedmann, M.; Kallipolitis, B.H. A small RNA controls expression of the chitinase ChiA in Listeria monocytogenes. PLoS ONE 2011, 6, e19019. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, J.K.; Nielsen, J.S.; Ebersbach, T.; Valentin-Hansen, P.; Sogaard-Andersen, L.; Kallipolitis, B.H. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA 2006, 12, 1383–1396. [Google Scholar] [CrossRef] [PubMed]
- Obana, N.; Shirahama, Y.; Abe, K.; Nakamura, K. Stabilization of Clostridium perfringens collagenase mRNA by VR-RNA-dependent cleavage in 5′-leader sequence. Mol. Microbiol. 2010, 77, 1416–1428. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Yaguchi, H.; Ohtani, K.; Banu, S.; Hayashi, H. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Mol. Microbiol. 2002, 43, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Obana, N.; Nomura, N.; Nakamura, K. Structural requirement in Clostridium perfringens collagenase mRNA 5′-leader sequence for translational induction through small RNA-mRNA base pairing. J. Bacteriol. 2013, 195, 2937–2946. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, K.; Bhowmik, S.K.; Hayashi, H.; Shimizu, T. Identification of a novel locus that regulates expression of toxin genes in Clostridium perfringens. FEMS Microbiol. Lett. 2002, 209, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Shioya, K.; Michaux, C.; Kuenne, C.; Hain, T.; Verneuil, N.; Budin-Verneuil, A.; Hartsch, T.; Hartke, A.; Giard, J.C. Genome-wide identification of small RNAs in the opportunistic pathogen Enterococcus faecalis v583. PLoS ONE 2011, 6, e23948. [Google Scholar] [CrossRef] [PubMed]
- Mangold, M.; Siller, M.; Roppenser, B.; Vlaminckx, B.J.; Penfound, T.A.; Klein, R.; Novak, R.; Novick, R.P.; Charpentier, E. Synthesis of Group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Mol. Microbiol. 2004, 53, 1515–1527. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ploplis, V.A.; French, E.L.; Boyle, M.D. Interaction between Group A streptococci and the plasmin(ogen) system promotes virulence in a mouse skin infection model. J. Infect. Dis. 1999, 179, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Patenge, N.; Billion, A.; Raasch, P.; Normann, J.; Wisniewska-Kucper, A.; Retey, J.; Boisguerin, V.; Hartsch, T.; Hain, T.; Kreikemeyer, B. Identification of novel growth phase- and media-dependent small non-coding RNAs in Streptococcus pyogenes M49 using intergenic tiling arrays. BMC Genom. 2012, 13, 550. [Google Scholar] [CrossRef] [PubMed]
- Perez, N.; Trevino, J.; Liu, Z.; Ho, S.C.; Babitzke, P.; Sumby, P. A genome-wide analysis of small regulatory RNAs in the human pathogen Group A Streptococcus. PLoS ONE 2009, 4, e7668. [Google Scholar] [CrossRef] [PubMed]
- Patenge, N.; Pappesch, R.; Khani, A.; Kreikemeyer, B. Genome-wide analyses of small non-coding RNAs in streptococci. Front. Genet. 2015, 6, 189. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Trevino, J.; Ramirez-Pena, E.; Sumby, P. The small regulatory RNA FasX controls pilus expression and adherence in the human bacterial pathogen Group A Streptococcus. Mol. Microbiol. 2012, 86, 140–154. [Google Scholar] [CrossRef] [PubMed]
- Sanderson-Smith, M.L.; de Oliveira, D.M.; Ranson, M.; McArthur, J.D. Bacterial plasminogen receptors: Mediators of a multifaceted relationship. J. Biomed. Biotechnol. 2012, 2012, 272148. [Google Scholar] [CrossRef] [PubMed]
- Syrovets, T.; Lunov, O.; Simmet, T. Plasmin as a proinflammatory cell activator. J. Leukoc. Biol. 2012, 92, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Podkaminski, D.; Vogel, J. Small RNAs promote mRNA stability to activate the synthesis of virulence factors. Mol. Microbiol. 2010, 78, 1327–1331. [Google Scholar] [CrossRef] [PubMed]
- Quigley, B.R.; Zahner, D.; Hatkoff, M.; Thanassi, D.G.; Scott, J.R. Linkage of T3 and Cpa pilins in the Streptococcus pyogenes M3 pilus. Mol. Microbiol. 2009, 72, 1379–1394. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.R.; Smoot, L.M.; Migliaccio, C.A.; Virtaneva, K.; Sturdevant, D.E.; Porcella, S.F.; Federle, M.J.; Adams, G.J.; Scott, J.R.; Musser, J.M. Virulence control in Group A Streptococcus by a two-component gene regulatory system: Global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. USA 2002, 99, 13855–13860. [Google Scholar] [CrossRef] [PubMed]
- Le Rhun, A.; Charpentier, E. Small RNAs in streptococci. RNA Biol. 2012, 9, 414–426. [Google Scholar] [CrossRef] [PubMed]
- Betschel, S.D.; Borgia, S.M.; Barg, N.L.; Low, D.E.; De Azavedo, J.C. Reduced virulence of Group A streptococcal Tn916 mutants that do not produce streptolysin S. Infect. Immun. 1998, 66, 1671–1679. [Google Scholar] [PubMed]
- Wilton, J.; Acebo, P.; Herranz, C.; Gomez, A.; Amblar, M. Small regulatory RNAs in Streptococcus pneumoniae: Discovery and biological functions. Front. Genet. 2015, 6, 126. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Shah, P.; Swiatlo, E.; Burgess, S.C.; Lawrence, M.L.; Nanduri, B. Identification of novel non-coding small RNAs from Streptococcus pneumoniae TIGR4 using high-resolution genome tiling arrays. BMC Genom. 2010, 11, 350. [Google Scholar] [CrossRef] [PubMed]
- Kowalko, J.E.; Sebert, M.E. The Streptococcus pneumoniae competence regulatory system influences respiratory tract colonization. Infect. Immun. 2008, 76, 3131–3140. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, Y. Diverse small non-coding RNAs in RNA interference pathways. Methods Mol. Biol. 2011, 764, 169–182. [Google Scholar] [PubMed]
- Acebo, P.; Martin-Galiano, A.J.; Navarro, S.; Zaballos, A.; Amblar, M. Identification of 88 regulatory small RNAs in the TIGR4 strain of the human pathogen Streptococcus pneumoniae. RNA 2012, 18, 530–546. [Google Scholar] [CrossRef] [PubMed]
- Marx, P.; Nuhn, M.; Kovacs, M.; Hakenbeck, R.; Bruckner, R. Identification of genes for small non-coding RNAs that belong to the regulon of the two-component regulatory system CiaRH in Streptococcus. BMC Genom. 2010, 11, 661. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, A.; Kovacs, M.; Hakenbeck, R.; Bruckner, R. Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: Five out of 15 promoters drive expression of small non-coding RNAs. Mol. Microbiol. 2007, 66, 110–126. [Google Scholar] [CrossRef] [PubMed]
- Blot, S.; Vandewoude, K.; Colardyn, F. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 2025–2026. [Google Scholar] [PubMed]
- Benito, Y.; Kolb, F.A.; Romby, P.; Lina, G.; Etienne, J.; Vandenesch, F. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA 2000, 6, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Mraheil, M.A.; Billion, A.; Mohamed, W.; Mukherjee, K.; Kuenne, C.; Pischimarov, J.; Krawitz, C.; Retey, J.; Hartsch, T.; Chakraborty, T.; et al. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res. 2011, 39, 4235–4248. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Arana, A.; Dussurget, O.; Nikitas, G.; Sesto, N.; Guet-Revillet, H.; Balestrino, D.; Loh, E.; Gripenland, J.; Tiensuu, T.; Vaitkevicius, K.; et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature 2009, 459, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Wurtzel, O.; Sesto, N.; Mellin, J.R.; Karunker, I.; Edelheit, S.; Becavin, C.; Archambaud, C.; Cossart, P.; Sorek, R. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol. Syst. Biol. 2012, 8, 583. [Google Scholar] [CrossRef] [PubMed]
- Vitreschak, A.G.; Rodionov, D.A.; Mironov, A.A.; Gelfand, M.S. Riboswitches: The oldest mechanism for the regulation of gene expression? Trends Genet. 2004, 20, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.S.; Lei, L.K.; Ebersbach, T.; Olsen, A.S.; Klitgaard, J.K.; Valentin-Hansen, P.; Kallipolitis, B.H. Defining a role for Hfq in gram-positive bacteria: Evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 2010, 38, 907–919. [Google Scholar] [CrossRef] [PubMed]
- Carter, G.P.; Cheung, J.K.; Larcombe, S.; Lyras, D. Regulation of toxin production in the pathogenic clostridia. Mol. Microbiol. 2014, 91, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Okumura, K.; Ohtani, K.; Hayashi, H.; Shimizu, T. Characterization of genes regulated directly by the VirR/VirS system in Clostridium perfringens. J. Bacteriol. 2008, 190, 7719–7727. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, K.; Hirakawa, H.; Paredes-Sabja, D.; Tashiro, K.; Kuhara, S.; Sarker, M.R.; Shimizu, T. Unique regulatory mechanism of sporulation and enterotoxin production in Clostridium perfringens. J. Bacteriol. 2013, 195, 2931–2936. [Google Scholar] [CrossRef] [PubMed]
- Kawsar, H.I.; Ohtani, K.; Okumura, K.; Hayashi, H.; Shimizu, T. Organization and transcriptional regulation of myo-inositol operon in Clostridium perfringens. FEMS Microbiol. Lett. 2004, 235, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, K.; Hirakawa, H.; Tashiro, K.; Yoshizawa, S.; Kuhara, S.; Shimizu, T. Identification of a two-component VirR/VirS regulon in Clostridium perfringens. Anaerobe 2010, 16, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Mraheil, M.A.; Billion, A.; Kuenne, C.; Pischimarov, J.; Kreikemeyer, B.; Engelmann, S.; Hartke, A.; Giard, J.C.; Rupnik, M.; Vorwerk, S.; et al. Comparative genome-wide analysis of small RNAs of major gram-positive pathogens: From identification to application. Microb. Biotechnol. 2010, 3, 658–676. [Google Scholar] [CrossRef] [PubMed]
- Michaux, C.; Hartke, A.; Martini, C.; Reiss, S.; Albrecht, D.; Budin-Verneuil, A.; Sanguinetti, M.; Engelmann, S.; Hain, T.; Verneuil, N.; et al. Involvement of Enterococcus faecalis small RNAs in stress response and virulence. Infect. Immun. 2014, 82, 3599–3611. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Singh, K.V.; Weinstock, G.M.; Murray, B.E. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 2001, 183, 3372–3382. [Google Scholar] [CrossRef] [PubMed]
- Verneuil, N.; Rince, A.; Sanguinetti, M.; Posteraro, B.; Fadda, G.; Auffray, Y.; Hartke, A.; Giard, J.C. Contribution of a PerR-like regulator to the oxidative-stress response and virulence of Enterococcus faecalis. Microbiology 2005, 151, 3997–4004. [Google Scholar] [CrossRef] [PubMed]
- Weaver, K.E.; Jensen, K.D.; Colwell, A.; Sriram, S.I. Functional analysis of the Enterococcus faecalis plasmid pAD1-encoded stability determinant par. Mol. Microbiol. 1996, 20, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Weaver, K.E. Emerging plasmid-encoded antisense RNA regulated systems. Curr. Opin. Microbiol. 2007, 10, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Geissmann, T.; Chevalier, C.; Cros, M.J.; Boisset, S.; Fechter, P.; Noirot, C.; Schrenzel, J.; Francois, P.; Vandenesch, F.; Gaspin, C.; et al. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 2009, 37, 7239–7257. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Emerson, N.; Gahan, C.G.; Hill, C. Identification and disruption of LisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J. Bacteriol. 1999, 181, 6840–6843. [Google Scholar] [PubMed]
- Levin, J.C.; Wessels, M.R. Identification of csrr/csrs, a genetic locus that regulates hyaluronic acid capsule synthesis in Group A Streptococcus. Mol. Microbiol. 1998, 30, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Churchward, G. The two faces of Janus: Virulence gene regulation by CovR/S in Group A streptococci. Mol. Microbiol. 2007, 64, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Mellin, J.R.; Cossart, P. Unexpected versatility in bacterial riboswitches. Trends Genet. 2015, 31, 150–156. [Google Scholar] [CrossRef] [PubMed]
- DebRoy, S.; Gebbie, M.; Ramesh, A.; Goodson, J.R.; Cruz, M.R.; van Hoof, A.; Winkler, W.C.; Garsin, D.A. Riboswitches. A riboswitch-containing sRNA controls gene expression by sequestration of a response regulator. Science 2014, 345, 937–940. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Vogel, J. Regulatory RNA in bacterial pathogens. Cell Host Microb. 2010, 8, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Lenz, D.H.; Mok, K.C.; Lilley, B.N.; Kulkarni, R.V.; Wingreen, N.S.; Bassler, B.L. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 2004, 118, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Tu, K.C.; Bassler, B.L. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007, 21, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Queck, S.Y.; Jameson-Lee, M.; Villaruz, A.E.; Bach, T.H.; Khan, B.A.; Sturdevant, D.E.; Ricklefs, S.M.; Li, M.; Otto, M. RNAIII-independent target gene control by the agr quorum-sensing system: Insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 2008, 32, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Autret, N.; Raynaud, C.; Dubail, I.; Berche, P.; Charbit, A. Identification of the agr locus of Listeria monocytogenes: Role in bacterial virulence. Infect. Immun. 2003, 71, 4463–4471. [Google Scholar] [CrossRef] [PubMed]
- Fozo, E.M.; Makarova, K.S.; Shabalina, S.A.; Yutin, N.; Koonin, E.V.; Storz, G. Abundance of type I toxin-antitoxin systems in bacteria: Searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010, 38, 3743–3759. [Google Scholar] [CrossRef] [PubMed]
- Sayed, N.; Nonin-Lecomte, S.; Rety, S.; Felden, B. Functional and structural insights of a Staphylococcus aureus apoptotic-like membrane peptide from a toxin-antitoxin module. J. Biol. Chem. 2012, 287, 43454–43463. [Google Scholar] [CrossRef] [PubMed]
- Aiba, H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007, 10, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Morita, T.; Maki, K.; Aiba, H. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005, 19, 2176–2186. [Google Scholar] [CrossRef] [PubMed]
- Kay, E.; Humair, B.; Denervaud, V.; Riedel, K.; Spahr, S.; Eberl, L.; Valverde, C.; Haas, D. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 6026–6033. [Google Scholar] [CrossRef] [PubMed]
- Mondragon, V.; Franco, B.; Jonas, K.; Suzuki, K.; Romeo, T.; Melefors, O.; Georgellis, D. pH-dependent activation of the BarA-UvrY two-component system in Escherichia coli. J. Bacteriol. 2006, 188, 8303–8306. [Google Scholar] [CrossRef] [PubMed]
- Barquist, L.; Vogel, J. Accelerating discovery and functional analysis of small RNAs with new technologies. Annu. Rev. Genet. 2015, 49, 367–394. [Google Scholar] [CrossRef] [PubMed]
- Brantl, S. Bacterial chromosome-encoded small regulatory RNAs. Future Microbiol. 2009, 4, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Ortega, A.D.; Quereda, J.J.; Pucciarelli, M.G.; Garcia-del Portillo, F. Non-coding RNA regulation in pathogenic bacteria located inside eukaryotic cells. Front. Cell Infect. Microbiol. 2014, 4, 162. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pitman, S.; Cho, K.H. The Mechanisms of Virulence Regulation by Small Noncoding RNAs in Low GC Gram-Positive Pathogens. Int. J. Mol. Sci. 2015, 16, 29797-29814. https://doi.org/10.3390/ijms161226194
Pitman S, Cho KH. The Mechanisms of Virulence Regulation by Small Noncoding RNAs in Low GC Gram-Positive Pathogens. International Journal of Molecular Sciences. 2015; 16(12):29797-29814. https://doi.org/10.3390/ijms161226194
Chicago/Turabian StylePitman, Stephanie, and Kyu Hong Cho. 2015. "The Mechanisms of Virulence Regulation by Small Noncoding RNAs in Low GC Gram-Positive Pathogens" International Journal of Molecular Sciences 16, no. 12: 29797-29814. https://doi.org/10.3390/ijms161226194






