PARP Inhibitor PJ34 Suppresses Osteogenic Differentiation in Mouse Mesenchymal Stem Cells by Modulating BMP-2 Signaling Pathway
Abstract
:1. Introduction
2. Results
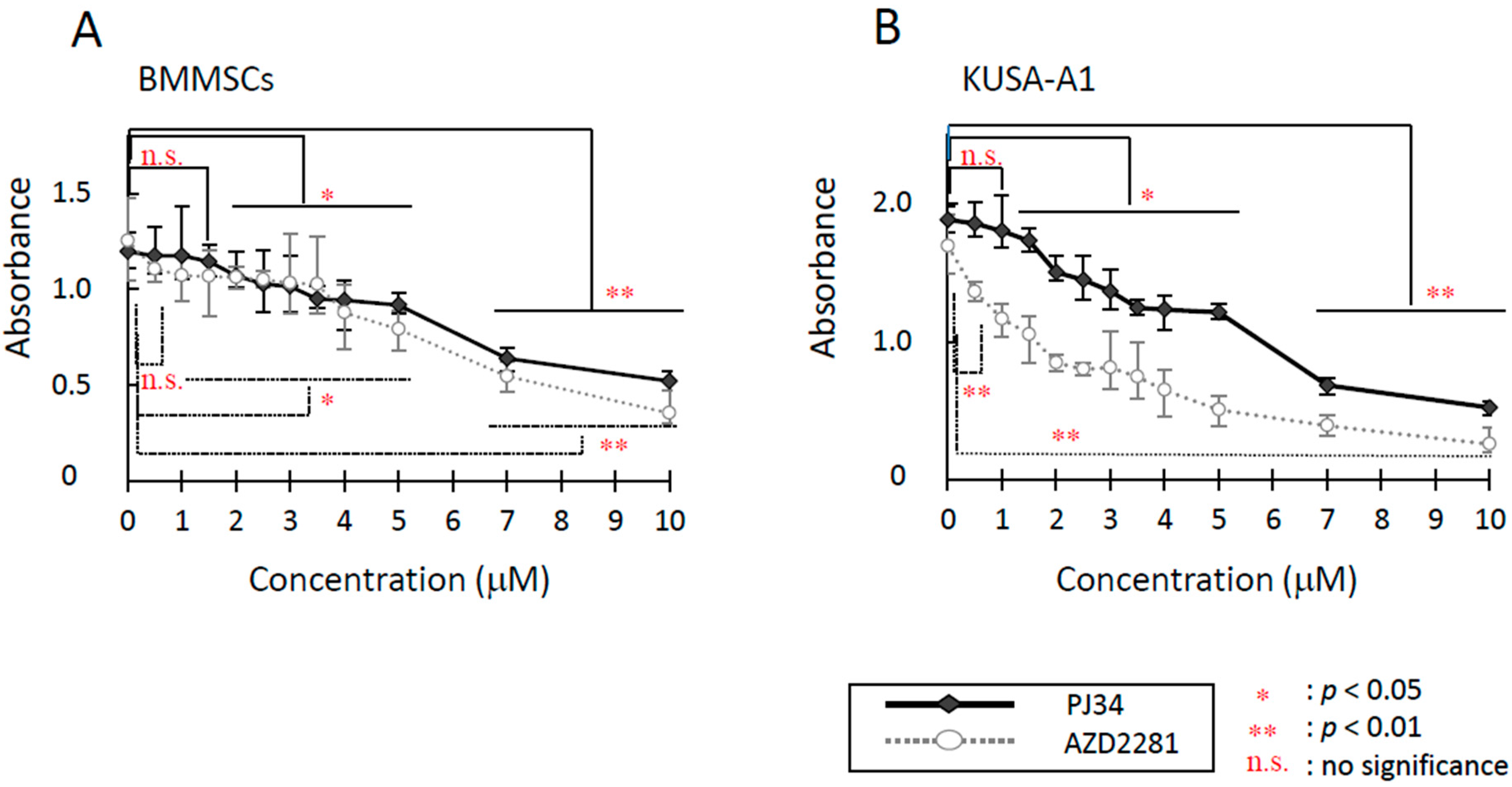
2.1. Cytotoxicity Evaluation
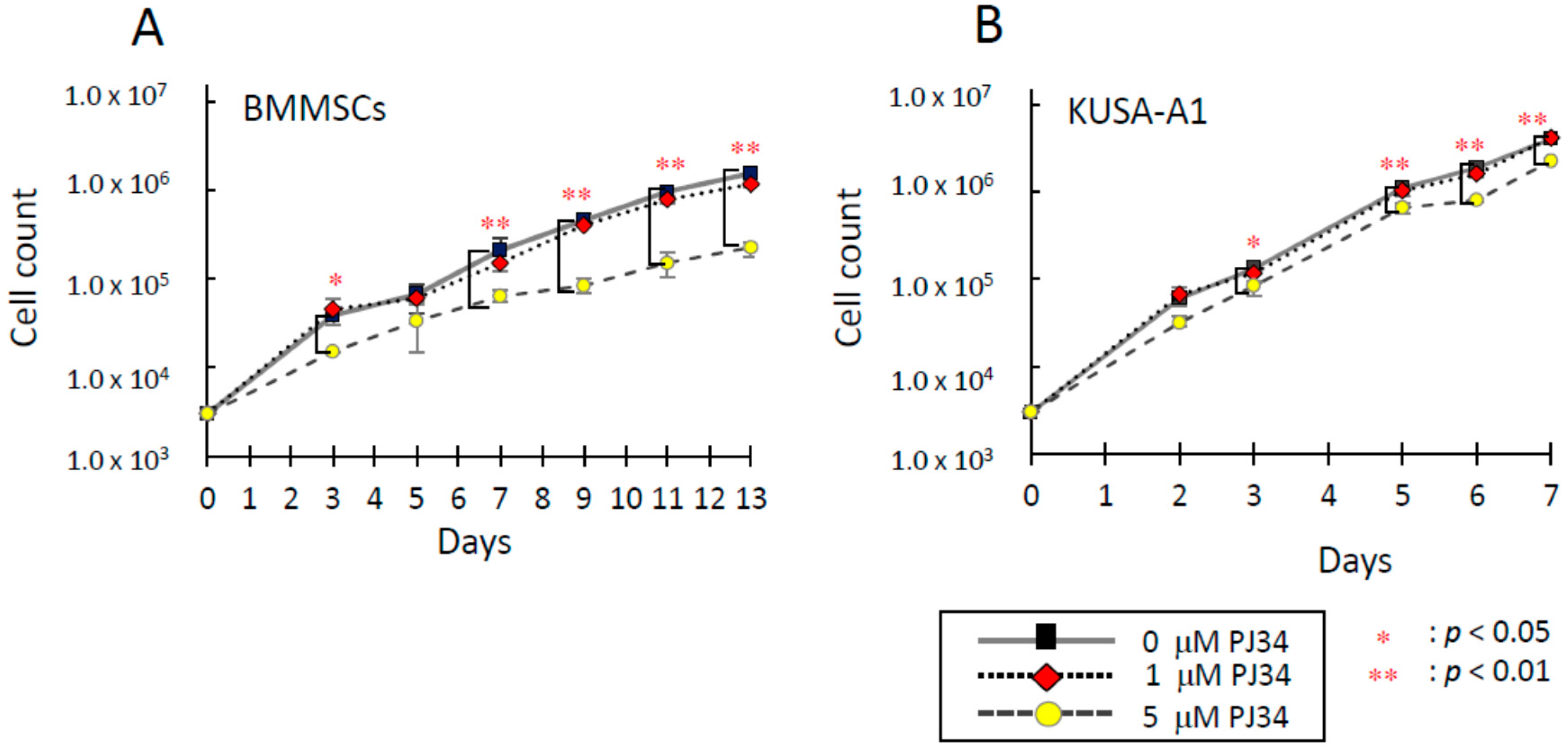
2.2. Effects of PJ34 on Cell Proliferation
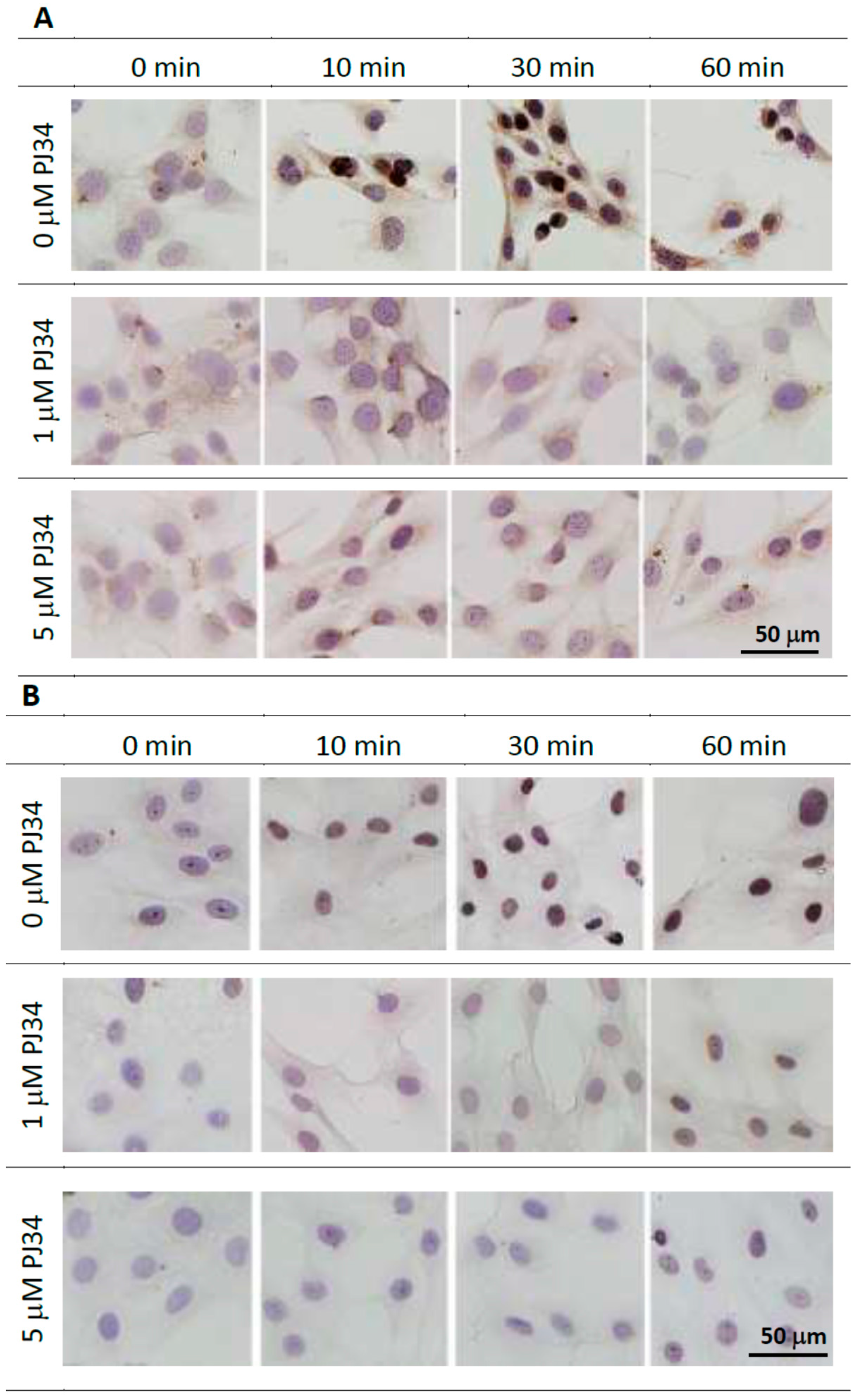
2.3. Effects of PJ34 on Poly(ADP-ribosyl)ation
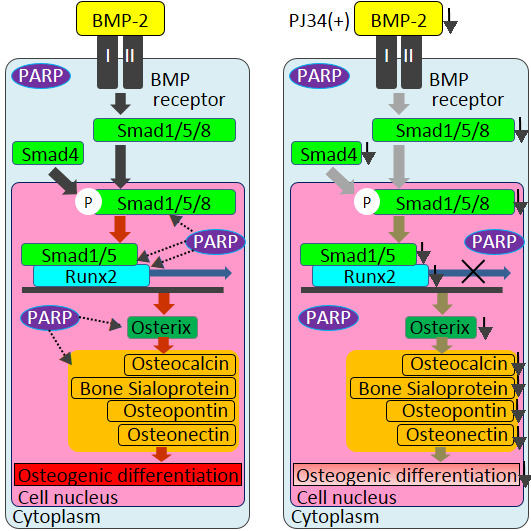
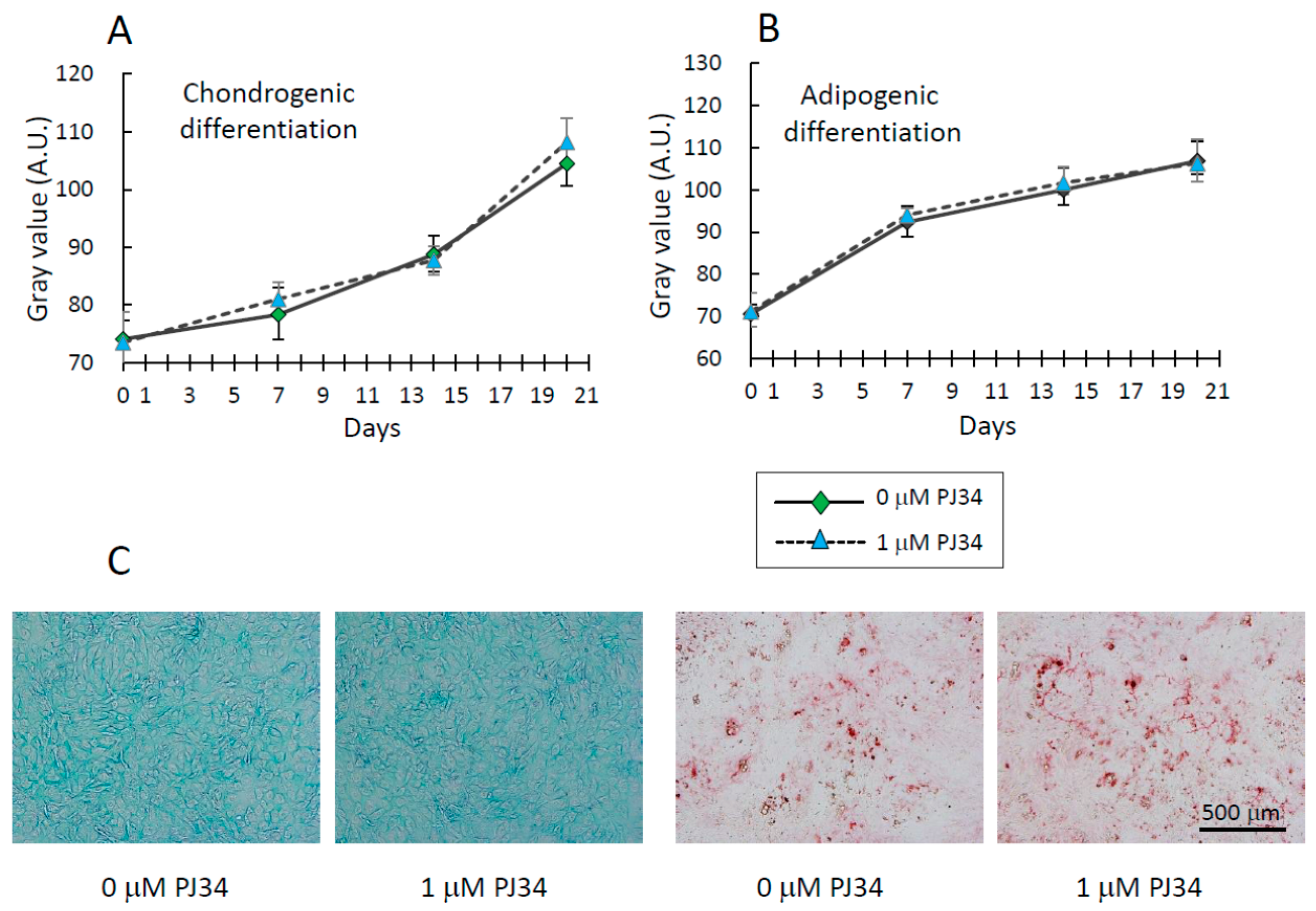
2.4. Effects of PJ34 on Osteogenic, Adipogenic and Chondrogenic Differentiation of MSCs
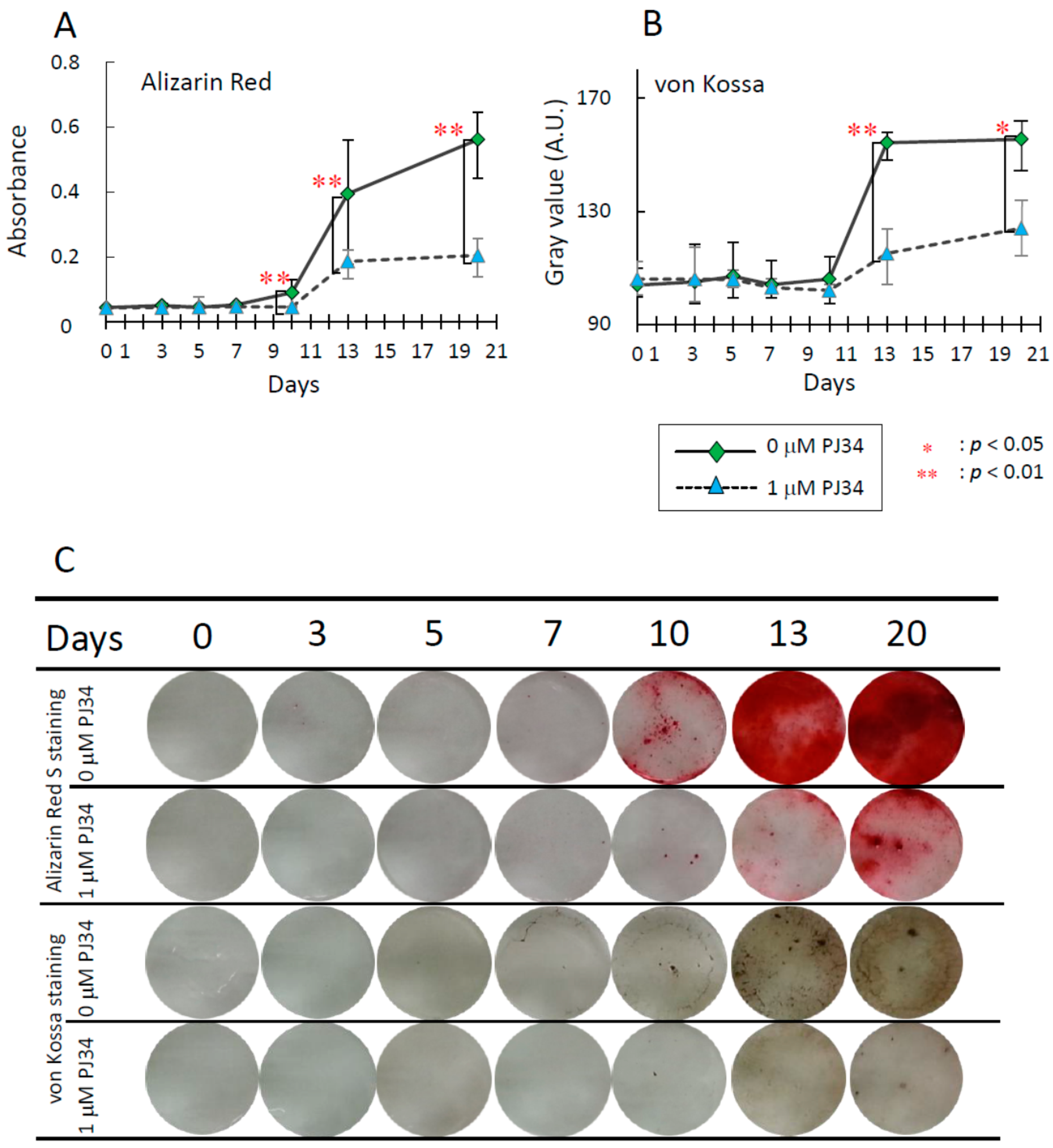


2.5. Effects of PJ34 on the Osteogenic Differentiation Markers
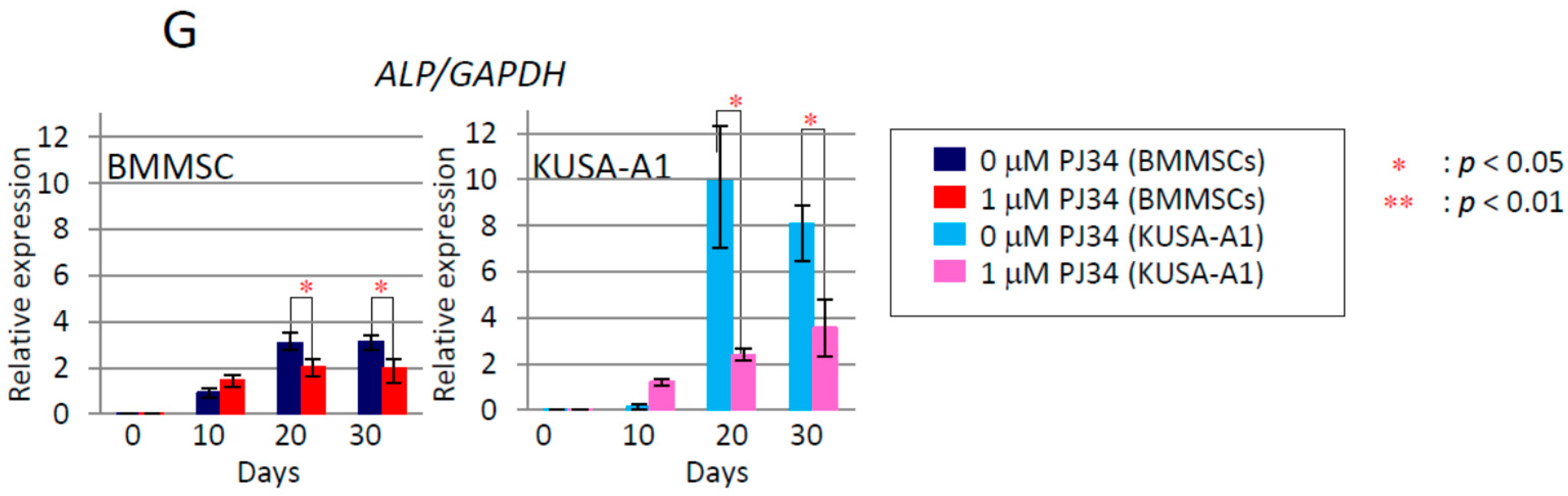
2.6. Effects of PJ34 on Osteogenic Differentiation Marker Protein Levels



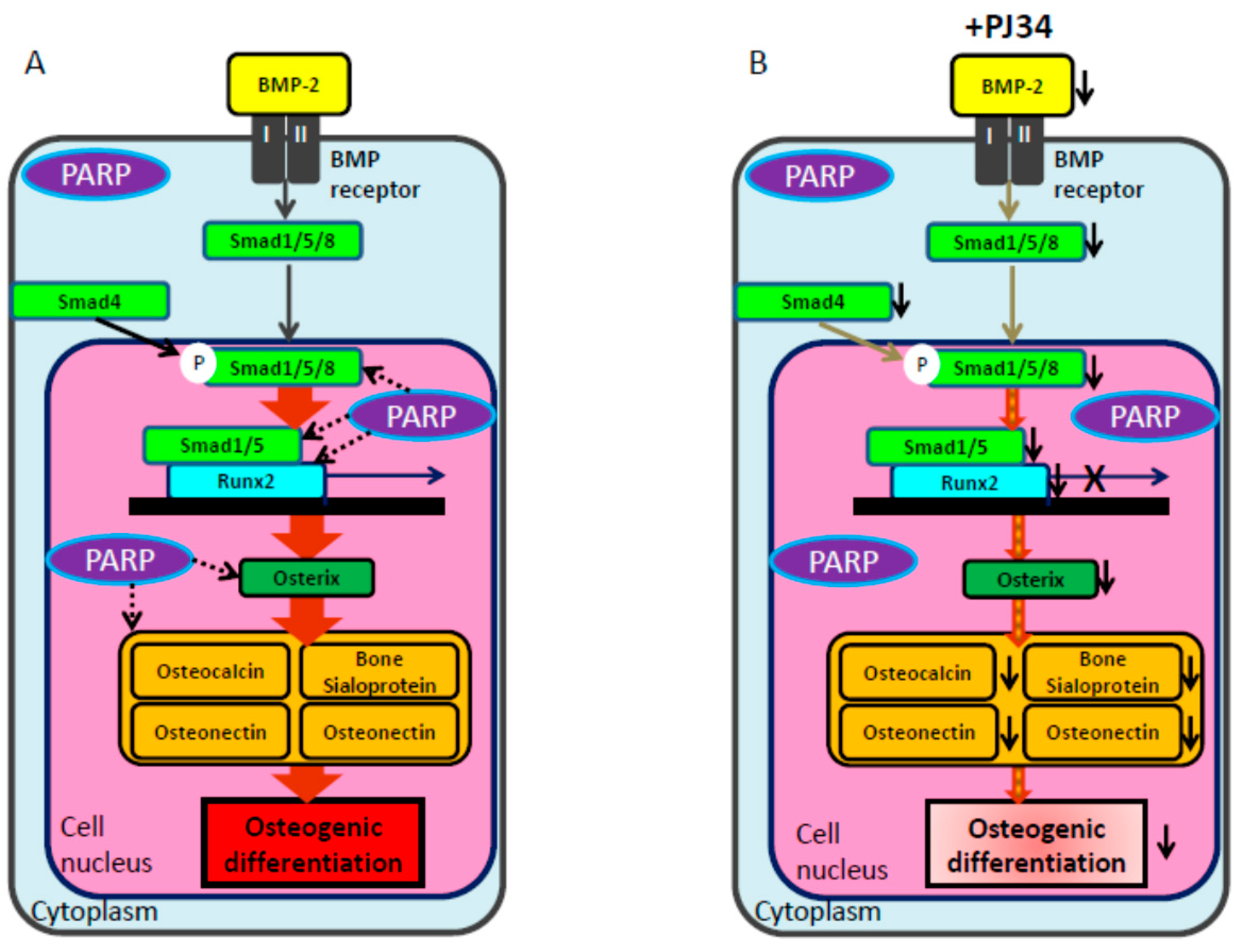
3. Discussion
4. Experimental Section
4.1. Culture of Mouse Bone Marrow-Derived Mesenchymal Stem Cells
4.2. MTT (Microculture Tetrazolium) Assay
4.3. Survival Assay
4.4. Proliferation Assay
4.5. Immunocytochemical Detection of Poly(ADP-ribose)
4.6. Osteogenic Cell Differentiation and Analysis
4.7. Chondrogenic Cell Differentiation and Analysis
4.8. Adipogenic Cell Differentiation and Analysis
4.9. Total RNA Extraction and Real-Time PCR
| Gene | Length | Forward Sequence (5′>3′) | Reverse Sequence (5′>3′) |
|---|---|---|---|
| Runx2 | 250 bp | CCACCACTCACTACCACACG | TCAGCGTCAACACCATCATT |
| Osterix | 373 bp | ACCAGGTCCAGGCAACACACCTAC | GCAGTCGCAGGTAGAACGCCCTGC |
| BMP2 | 154 bp | GGGACCCGCTGTCTTCTAGT | TCAACTCAAATTCGCTGAGGAC |
| Osteocalcin | 302 bp | AGACAAGTCCCACACAGCAG | GGCGGTCTTCAAGCCATACT |
| Bone sialoproiten | 312 bp | CTGAAGCAGGTGCAGAAGGA | TCTGACCCTCGTAGCCTTCA |
| Osteopontin | 354 bp | CTGGCAGCTCAGAGGAGAAG | GGACATCGACTGTAGGGACG |
| ALP | 211 bp | TAACACCAACGCTCAGGTCC | GTGGTTCACCCGAGTGGTAG |
| Smad1 | 97 bp | ACCTGTGGCTTCCGTCTC | ATCGTGGCTCCTTCGTC |
| Smad4 | 145 bp | CAGCACTACCACCTGGACTGGA | CTGGAATGCAAGCTCATTGTGAA |
| Smad5 | 197 bp | AAGTAGATTCTGCCTGGGATT | AGACGGTGGTGGGATGG |
| Smad8 | 169 bp | ATCCCTGGCAATCTGTA | CCCTGGCTGTCCTGTAA |
| PARP-1 | 72 bp | GGAAAGGGATCTACTTTGCCG | TCGGGTCTCCCTGAGATGTG |
| GAPDH | 240 bp | TGATGACATCAAGAAGGTGGTGAAG | TCCTTGGAGGCCATGTAGGCCAT |
4.10. Western Blot Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Huang, W.; Yang, S.; Shao, J.; Li, Y.P. Signaling and transcriptional regulation in osteoblast commitment and differentiation. Front. Biosci. 2007, 12, 3068–3092. [Google Scholar] [CrossRef] [PubMed]
- Fakhry, M.; Hamade, E.; Badran, B.; Buchet, R.; Magne, D. Molecular mechanisms of mesenchymal stem cell differentiation towards osteoblasts. World J. Stem Cells 2013, 5, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Deng, C.; Li, Y.P. TGF-β and BMP signaling in osteoblast differentiation and bone formation. Int. J. Biol. Sci. 2012, 8, 272–288. [Google Scholar] [CrossRef] [PubMed]
- Phimphilai, M.; Zhao, Z.; Boules, H.; Roca, H.; Franceschi, R.T. BMP signaling is required for RUNX2-dependent induction of the osteoblast phenotype. J. Bone Miner. Res. 2006, 21, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Malanga, M.; Pleschke, J.M.; Kleczkowska, H.E.; Althaus, F.R. Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J. Biol. Chem. 1998, 273, 11839–11843. [Google Scholar] [CrossRef] [PubMed]
- Pleschke, J.M.; Kleczkowska, H.E.; Strohm, M.; Althaus, F.R. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000, 275, 40974–40980. [Google Scholar] [CrossRef] [PubMed]
- Morales, J.; Li, L.; Fattah, F.J.; Dong, Y.; Bey, E.A.; Patel, M.; Gao, J.; Boothman, D.A. Review of poly(ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit. Rev. Eukaryot. Gene Expr. 2014, 24, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Seimiya, H. The telomeric PARP, tankyrases, as targets for cancer therapy. Br. J. Cancer 2006, 94, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Masutani, M.; Fujimori, H. Poly(ADP-ribosyl)ation in carcinogenesis. Mol. Asp. Med. 2013, 34, 1202–1216. [Google Scholar] [CrossRef] [PubMed]
- Masutani, M.; Nozaki, T.; Nishiyama, E.; Shimokawa, T.; Tachi, Y.; Suzuki, H.; Nakagama, H.; Wakabayashi, K.; Sugimura, T. Function of poly(ADP-ribose) polymerase in response to DNA damage: Gene-disruption study in mice. Mol. Cell. Biochem. 1999, 193, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Tahara, M.; Inoue, T.; Sato, F.; Miyakura, Y.; Horie, H.; Yasuda, Y.; Fujii, H.; Kotake, K.; Sugano, K. The use of olaparib (AZD2281) potentiates SN-38 cytotoxicity in colon cancer cells by indirect inhibition of Rad51-mediated repair of DNA double-strand breaks. Mol. Cancer Ther. 2014, 5, 1170–1180. [Google Scholar] [CrossRef] [PubMed]
- Ratnam, K.; Low, J.A. Current development of clinical inhibitors of poly(ADP-ribose) polymerase in oncology. Clin. Cancer Res. 2007, 13, 1383–1388. [Google Scholar] [CrossRef] [PubMed]
- Del Conte, G.; Sessa, C.; von Moos, R.; Vigano, L.; Digena, T.; Locatelli, A.; Gallerani, E.; Fasolo, A.; Tessari, A.; Cathomas, R.; et al. Phase I study of olaparib in combination with liposomal doxorubicin in patients with advanced solid tumours. Br. J. Cancer 2014, 111, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.F.; Tolaney, S.M.; Birrer, M.; Fleming, G.F.; Buss, M.K.; Dahlberg, S.E.; Lee, H.; Whalen, C.; Tyburski, K.; Winer, E.; et al. A phase I trial of the poly(ADP-ribose) polymerase inhibitor olaparib (AZD2281) in combination with the anti-angiogenic cediranib (AZD2171) in recurrent epithelial ovarian or triple-negative breast cancer. Eur. J. Cancer 2013, 49, 2972–2978. [Google Scholar] [CrossRef] [PubMed]
- Steffen, J.D.; Brody, J.R.; Armen, R.S.; Pascal, J.M. Structural implications for selective targeting of PARPS. Front. Oncol. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Fong, P.C.; Boss, D.S.; Yap, T.A.; Tutt, A.; Wu, P.; Mergui-Roelvink, M.; Mortimer, P.; Swaisland, H.; Lau, A.; O’Connor, M.J.; et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009, 361, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Bai, P.; Houten, S.M.; Huber, A.; Schreiber, V.; Watanabe, M.; Kiss, B.; de Murcia, G.; Auwerx, J.; Menissier-de Murcia, J. Poly(ADP-ribose) polymerase-2 controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/peroxisome proliferator-activated receptor-gamma heterodimer. J. Biol. Chem. 2007, 282, 37738–37746. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.; He, X.; Farmer, P.; Boden, S.; Kozlowski, M.; Rubin, J.; Nanes, M.S. Inhibition of osteoblast differentiation by tumor necrosis factor-α. Endocrinology 2000, 141, 3956–3964. [Google Scholar] [PubMed]
- Majewski, P.M.; Thurston, R.D.; Ramalingam, R.; Kiela, P.R.; Ghishan, F.K. Cooperative role of NF-κB and poly(ADP-ribose) polymerase 1 (PARP-1) in the TNF-induced inhibition of PHEX expression in osteoblasts. J. Biol. Chem. 2010, 285, 34828–34838. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Chen, L.L.; Kuo, P.Y.; Chang, J.L.; Wang, Y.J.; Hung, S.C. Apoptosis in chondrogenesis of human mesenchymal stem cells: Effect of serum and medium supplements. Apoptosis 2010, 15, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, A.; Shindo, Y.; Gomei, Y.; Mori, T.; Uyama, T.; Umezawa, A. Cell separation between mesenchymal progenitor cells through porous polymeric membranes. J. Biomed. Mater. Res. B Appl. Biomater. 2005, 74, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Mohammadian, M.; Shamsasenjan, K.; Lotfi Nezhad, P.; Talebi, M.; Jahedi, M.; Nickkhah, H.; Minayi, N.; Movassagh Pour, A. Mesenchymal stem cells: New aspect in cell-based regenerative therapy. Adv. Pharm. Bull. 2013, 3, 433–437. [Google Scholar] [PubMed]
- Zhang, Y.; Khan, D.; Delling, J.; Tobiasch, E. Mechanisms underlying the osteo- and adipo-differentiation of human mesenchymal stem cells. Sci. World J. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Peralta-Leal, A.; Rodriguez-Vargas, J.M.; Aguilar-Quesada, R.; Rodriguez, M.I.; Linares, J.L.; de Almodovar, M.R.; Oliver, F.J. Parp inhibitors: New partners in the therapy of cancer and inflammatory diseases. Free Radic. Biol. Med. 2009, 47, 13–26. [Google Scholar] [CrossRef] [PubMed]
- De Blasio, A.; Musmeci, M.T.; Giuliano, M.; Lauricella, M.; Emanuele, S.; D’Anneo, A.; Vassallo, B.; Tesoriere, G.; Vento, R. The effect of 3-aminobenzamide, inhibitor of poly(ADP-ribose) polymerase, on human osteosarcoma cells. Int. J. Oncol. 2003, 23, 1521–1528. [Google Scholar] [CrossRef] [PubMed]
- Robaszkiewicz, A.; Erdelyi, K.; Kovacs, K.; Kovacs, I.; Bai, P.; Rajnavolgyi, E.; Virag, L. Hydrogen peroxide-induced poly(ADP-ribosyl)ation regulates osteogenic differentiation-associated cell death. Free Radic. Biol. Med. 2012, 53, 1552–1564. [Google Scholar] [CrossRef] [PubMed]
- Madison, D.L.; Stauffer, D.; Lundblad, J.R. The PARP inhibitor PJ34 causes a PARP1-independent, p21 dependent mitotic arrest. DNA Repair 2011, 10, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Jouan-Lanhouet, S.; Arshad, M.I.; Piquet-Pellorce, C.; Martin-Chouly, C.; Le Moigne-Muller, G.; Van Herreweghe, F.; Takahashi, N.; Sergent, O.; Lagadic-Gossmann, D.; Vandenabeele, P.; et al. Trail induces necroptosis involving RIPK1/RIPK3-dependent PARP-1 activation. Cell Death Differ. 2012, 19, 2003–2014. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R. Parsing the phrase “all in for Axin”—Wnt pathway targets in cancer. Cancer Cell. 2009, 16, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Nanta, R.; Kumar, D.; Meeker, D.; Rodova, M.; Van Veldhuizen, P.J.; Shankar, S.; Srivastava, R.K. NVP-LDE-225 (Erismodegib) inhibits epithelial-mesenchymal transition and human prostate cancer stem cell growth in NOD/SCID IL2Rγ null mice by regulating Bmi-1 and microRNA-128. Oncogenesis 2013, 2. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, W. New molecular insights into osteosarcoma targeted therapy. Curr. Opin. Oncol. 2013, 25, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Lonn, P.; van der Heide, L.P.; Dahl, M.; Hellman, U.; Heldin, C.H.; Moustakas, A. PARP-1 attenuates Smad-mediated transcription. Mol. Cell 2010, 40, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Miura, M.; Miura, Y.; Padilla-Nash, H.M.; Molinolo, A.A.; Fu, B.; Patel, V.; Seo, B.M.; Sonoyama, W.; Zheng, J.J.; Baker, C.C.; et al. Accumulated chromosomal instability in murine bone marrow mesenchymal stem cells leads to malignant transformation. Stem Cells 2006, 24, 1095–1103. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhao, L.; Liu, J.; Liu, A.L.; Zeng, W.S.; Luo, S.Q.; Bai, X.C. Hydrogen peroxide induces G2 cell cycle arrest and inhibits cell proliferation in osteoblasts. Anat. Rec. 2009, 292, 1107–1113. [Google Scholar] [CrossRef] [PubMed]
- Burkle, A.; Chen, G.; Kupper, J.H.; Grube, K.; Zeller, W.J. Increased poly(ADP-ribosyl)ation in intact cells by cisplatin treatment. Carcinogenesis 1993, 14, 559–561. [Google Scholar] [CrossRef] [PubMed]
- Sila-Asna, M.; Bunyaratvej, A.; Maeda, S.; Kitaguchi, H.; Bunyaratavej, N. Osteoblast differentiation and bone formation gene expression in strontium-inducing bone marrow mesenchymal stem cell. Kobe J. Med. Sci. 2007, 53, 25–35. [Google Scholar] [PubMed]
- Satomura, K.; Tobiume, S.; Tokuyama, R.; Yamasaki, Y.; Kudoh, K.; Maeda, E.; Nagayama, M. Melatonin at pharmacological doses enhances human osteoblastic differentiation in vitro and promotes mouse cortical bone formation in vivo. J. Pineal Res. 2007, 42, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Bouffi, C.; Thomas, O.; Bony, C.; Giteau, A.; Venier-Julienne, M.C.; Jorgensen, C.; Montero-Menei, C.; Noel, D. The role of pharmacologically active microcarriers releasing TGF-β3 in cartilage formation in vivo by mesenchymal stem cells. Biomaterials 2010, 31, 6485–6493. [Google Scholar] [PubMed]
- Cunha, M.C.; Lima Fda, S.; Vinolo, M.A.; Hastreiter, A.; Curi, R.; Borelli, P.; Fock, R.A. Protein malnutrition induces bone marrow mesenchymal stem cells commitment to adipogenic differentiation leading to hematopoietic failure. PLoS ONE 2013, 8, e58872. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kishi, Y.; Fujihara, H.; Kawaguchi, K.; Yamada, H.; Nakayama, R.; Yamamoto, N.; Fujihara, Y.; Hamada, Y.; Satomura, K.; Masutani, M. PARP Inhibitor PJ34 Suppresses Osteogenic Differentiation in Mouse Mesenchymal Stem Cells by Modulating BMP-2 Signaling Pathway. Int. J. Mol. Sci. 2015, 16, 24820-24838. https://doi.org/10.3390/ijms161024820
Kishi Y, Fujihara H, Kawaguchi K, Yamada H, Nakayama R, Yamamoto N, Fujihara Y, Hamada Y, Satomura K, Masutani M. PARP Inhibitor PJ34 Suppresses Osteogenic Differentiation in Mouse Mesenchymal Stem Cells by Modulating BMP-2 Signaling Pathway. International Journal of Molecular Sciences. 2015; 16(10):24820-24838. https://doi.org/10.3390/ijms161024820
Chicago/Turabian StyleKishi, Yuta, Hisako Fujihara, Koji Kawaguchi, Hiroyuki Yamada, Ryoko Nakayama, Nanami Yamamoto, Yuko Fujihara, Yoshiki Hamada, Kazuhito Satomura, and Mitsuko Masutani. 2015. "PARP Inhibitor PJ34 Suppresses Osteogenic Differentiation in Mouse Mesenchymal Stem Cells by Modulating BMP-2 Signaling Pathway" International Journal of Molecular Sciences 16, no. 10: 24820-24838. https://doi.org/10.3390/ijms161024820