Experimental Confirmation of a Whole Set of tRNA Molecules in Two Archaeal Species
Abstract
:1. Introduction
2. Confirmation of a Whole Set of tRNA Molecules
2.1. The Whole Set of tRNA Molecules in Aeropyrum pernix K1
2.1.1. Introduction and Features of tRNAs in A. pernix K1
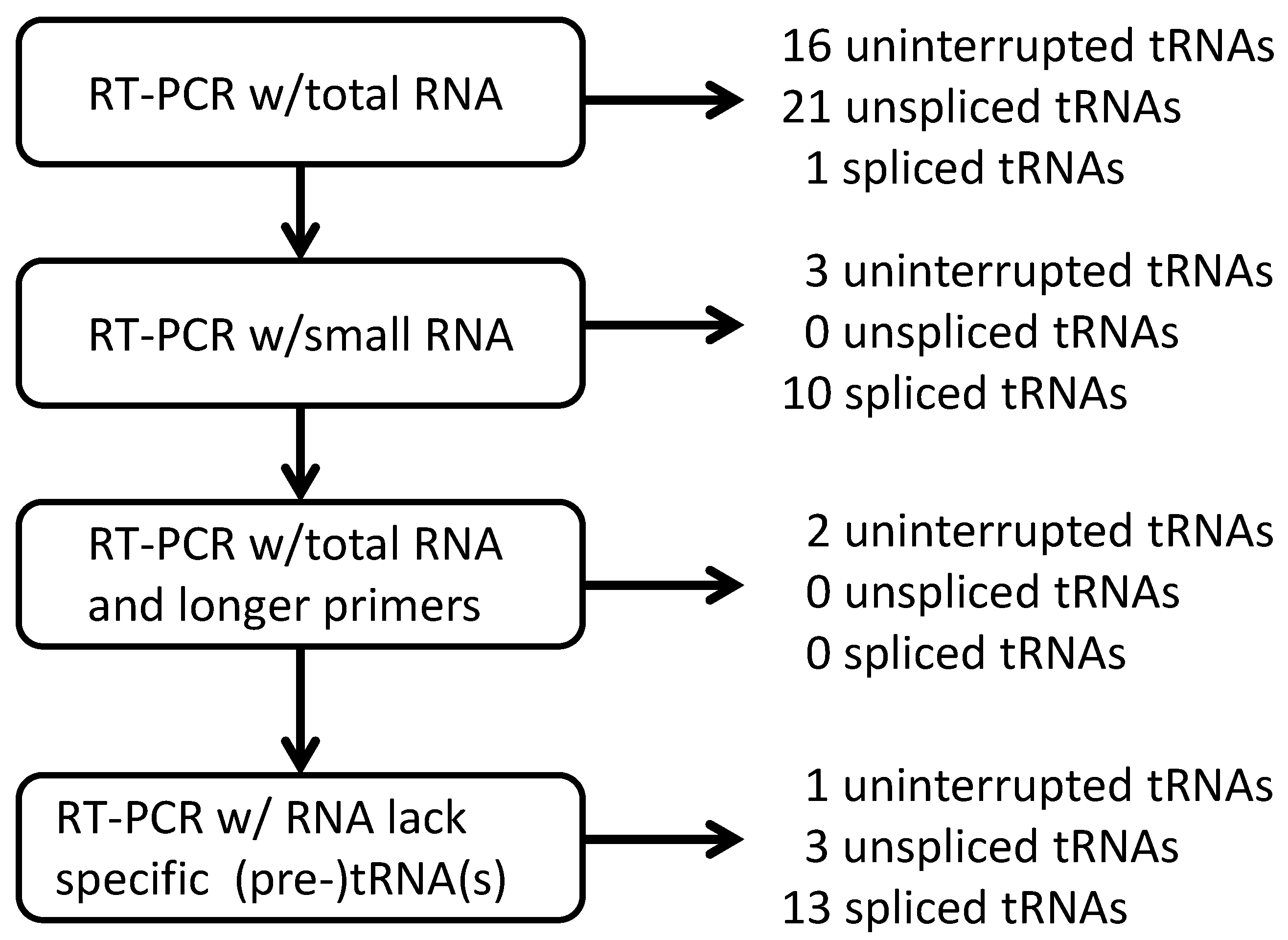
2.1.2. Strategies for Detection of RNA Molecules
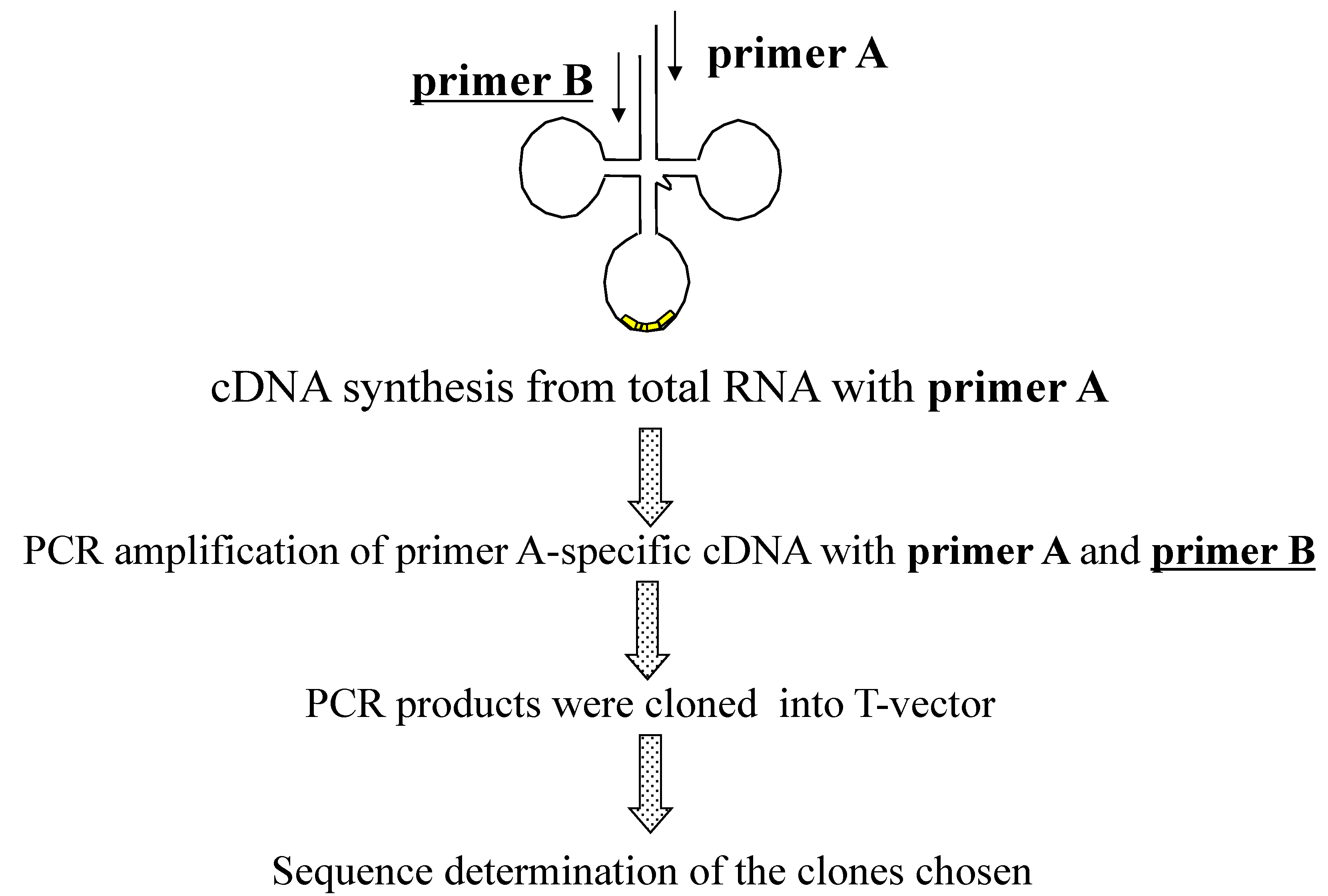
2.1.3. Isolation of cDNA of tRNA Molecules from Total RNA
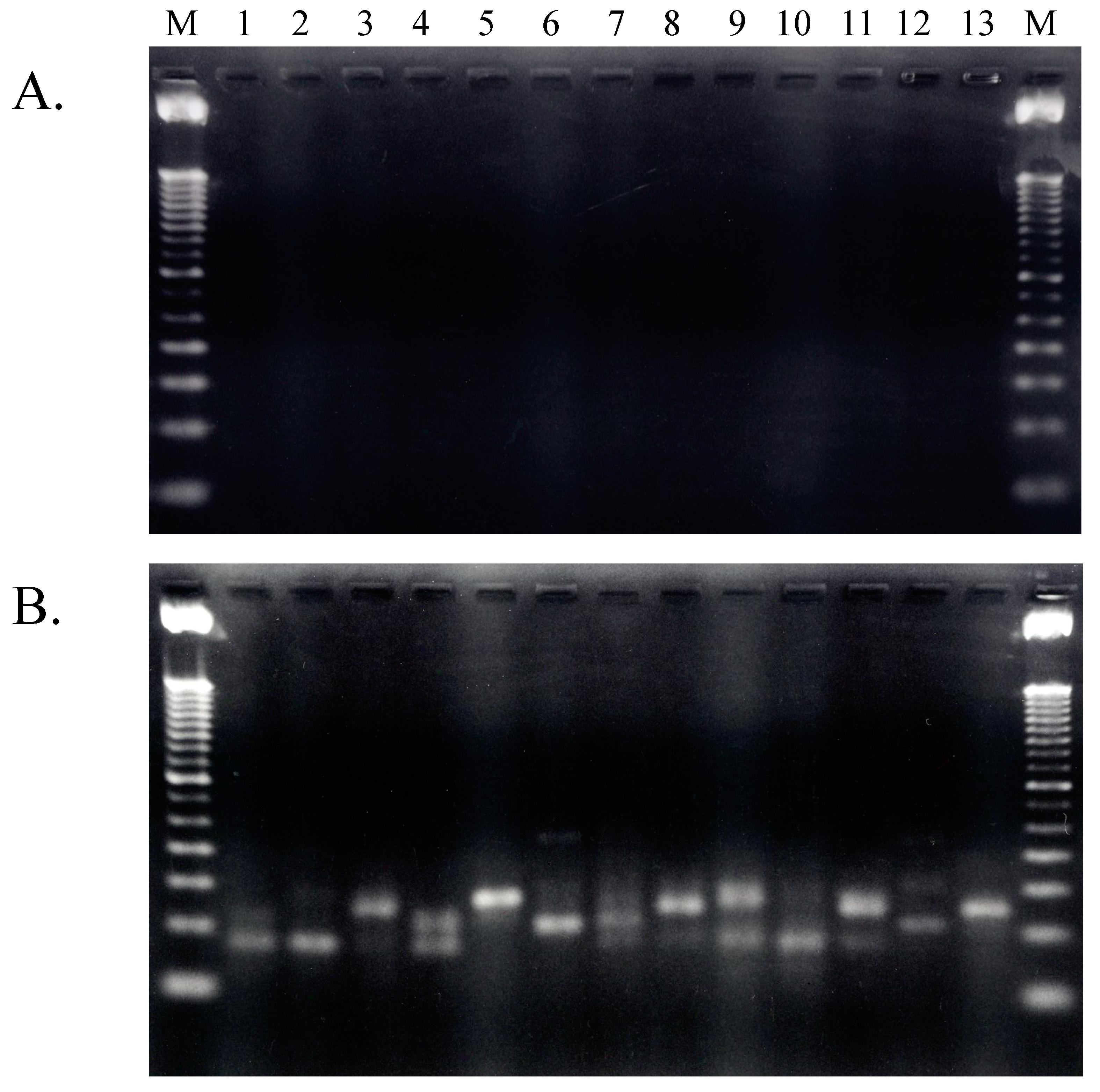
2.1.4. Isolation of tRNA Molecules from Small-Sized RNA and Species-Specific Concentrated RNA
| tRNA Species | un | sp | tRNA Species | un | sp | tRNA Species | un | sp | tRNA Species | un | sp |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phe(AAA) | - | - | Ser(AGA) | - | - | Tyr(AUA) | - | - | Cys(ACA) | - | - |
| Phe(GAA) | - | s | Ser(GGA) | - | s | Tyr(GUA) | t | s | Cys(GCA) | t | t |
| Leu(UAA) | - | t | Ser(UGA) | - | x | End(UUA) | - | - | End(UCA) | - | - |
| Leu(CAA) | - | t | Ser(CGA) | t | x | End(CUA) | - | - | Trp(CCA) * | t | c |
| Leu(AAG) | - | - | Pro(AGG) | - | - | His(AUG) | - | - | Arg(ACG) | - | - |
| Leu(GAG) | - | s | Pro(GGG) | t | t | His(GUG) | - | t | Arg(GCG) | - | t |
| Leu(UAG) | - | t | Pro(UGG) | - | t | Gln(UUG) | - | t | Arg(UCG) | - | t |
| Leu(CAG) | - | t | Pro(CGG) | t | t | Gln(CUG) | - | t | Arg(CCG) | - | t |
| Ile(AAU) | - | - | Thr(AGU) | - | - | Asn(AUU) | - | - | Ser(AGU) | - | - |
| Ile(GAU) | - | t | Thr(GGU) | - | t | Asn(GUU) | - | t | Ser(GGU) | - | t |
| Ile(UAU) | - | - | Thr(UGU) + | t, x | t, x | Lys(UUU) | t | t | Arg(UCU) | t | t |
| Met(CAU) # | t | t | Thr(CGU) | x | t | Lys(CUU) | t | t | Arg(CCU) | - | t |
| Val(AAC) | - | - | Ala(AGC) | - | - | Asp(AUC) | - | - | Gly(ACC) | - | - |
| Val(GAC) | - | t | Ala(GGC) | - | t | Asp(GUC) * | t | t | Gly(GCC) | - | t |
| Val(UAC) | - | t | Ala(UGC) | - | t | Glu(UUC) | - | t | Gly(UCC) | - | t |
| Val(CAC) | - | t | Ala(CGC) | - | t | Glu(CUC) | - | t | Gly(UCC) | - | t |
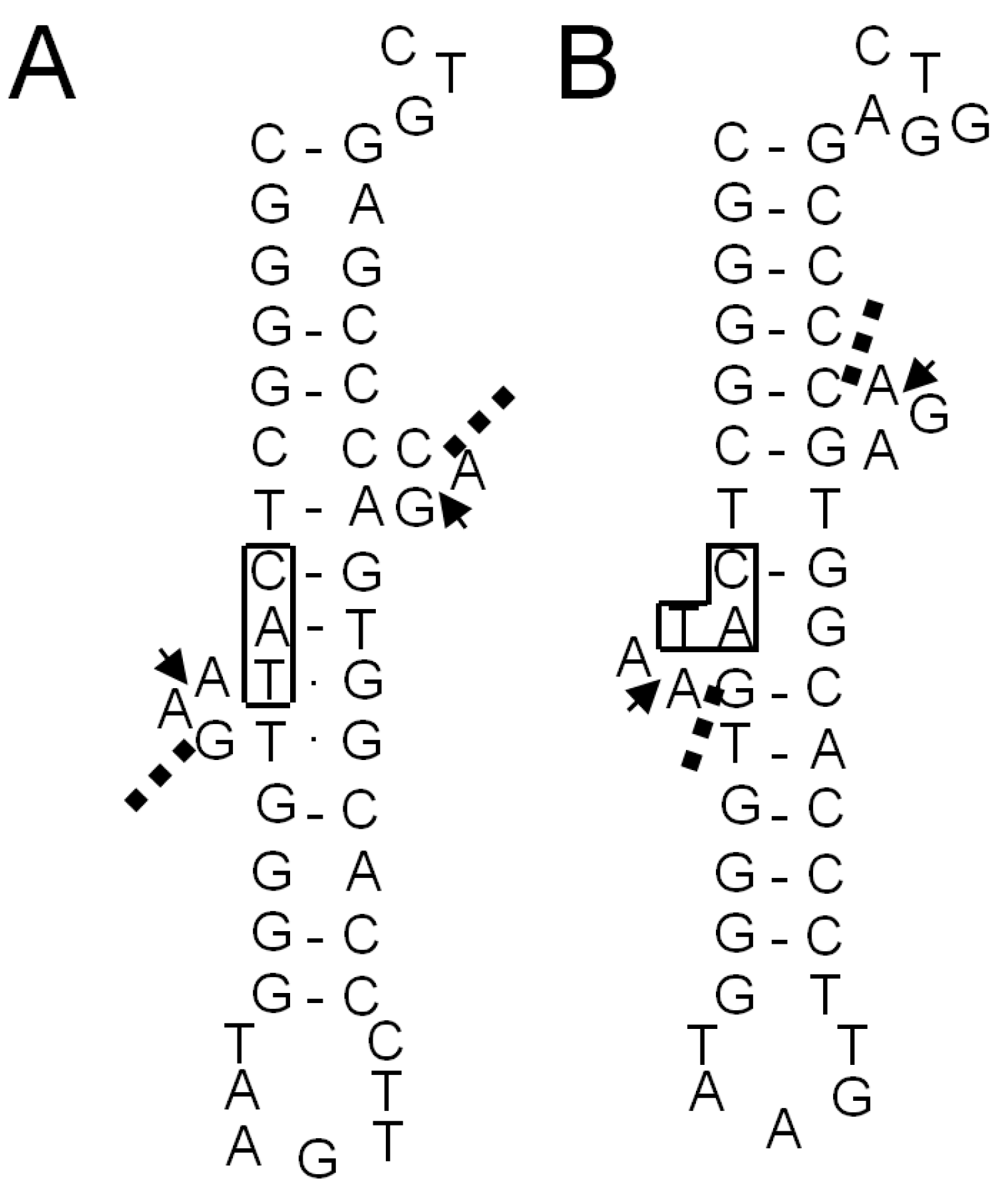
2.1.5. Confirmation of the Actual Splicing Patterns for Each Interrupted A. pernix tRNA Molecule
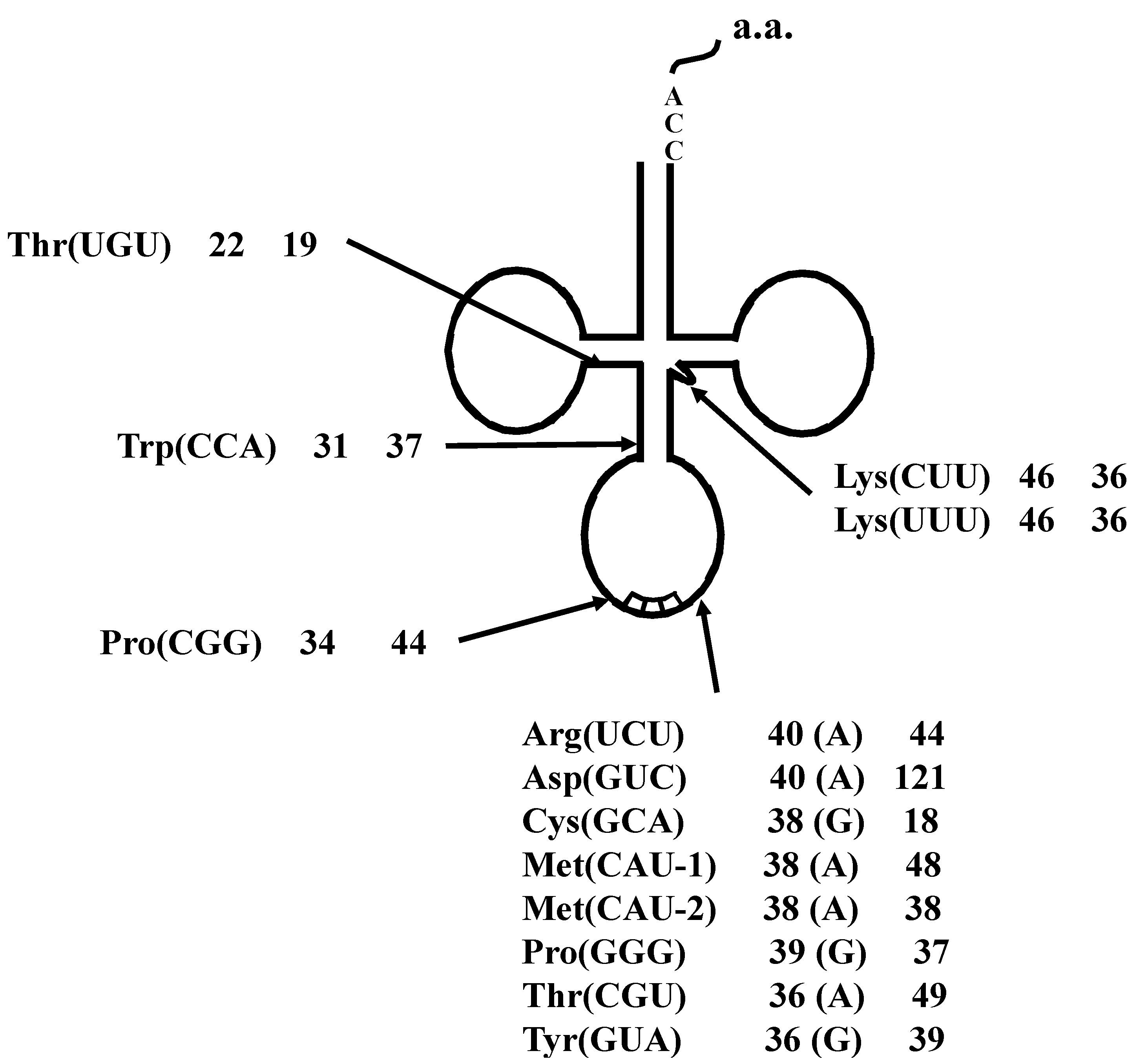
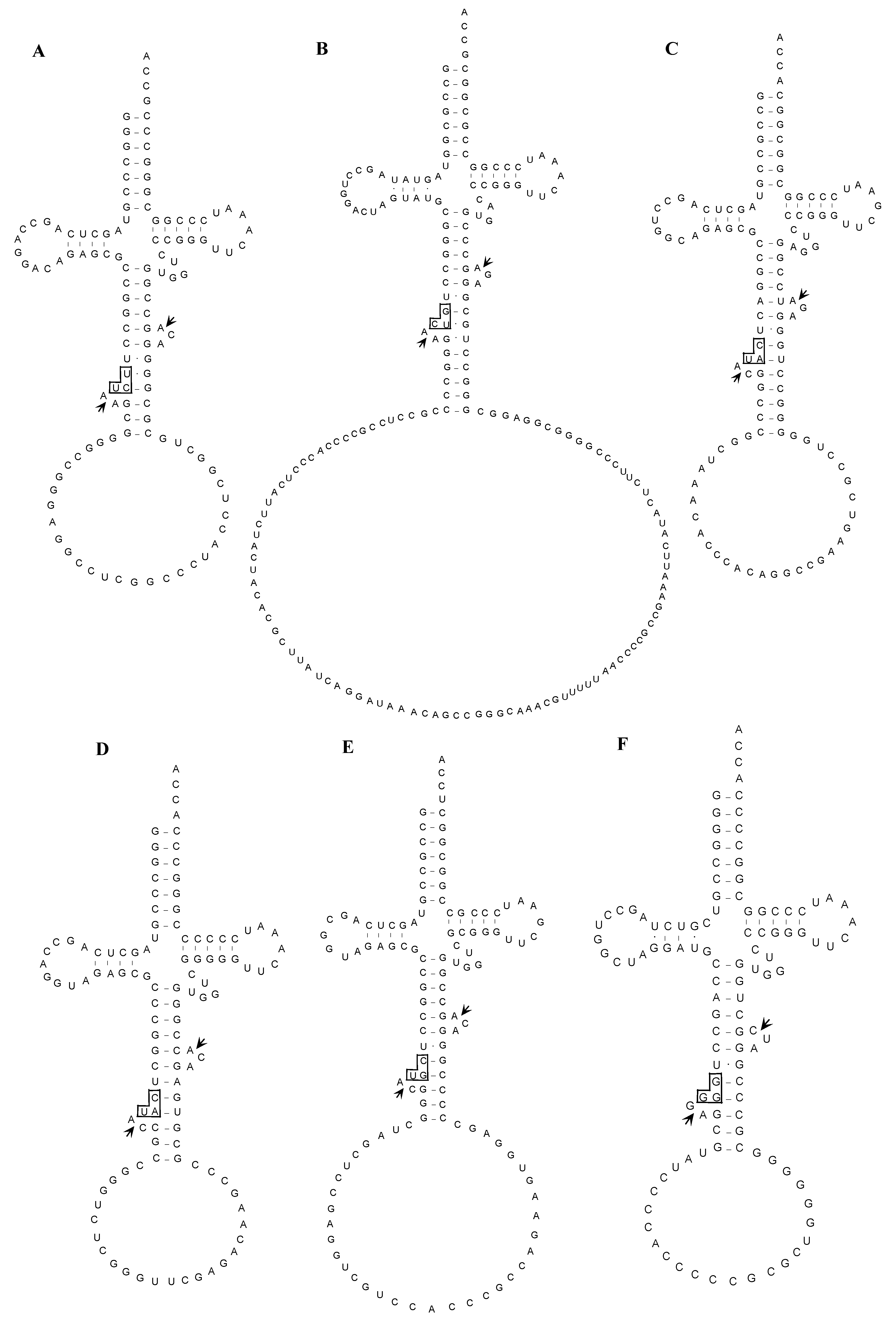
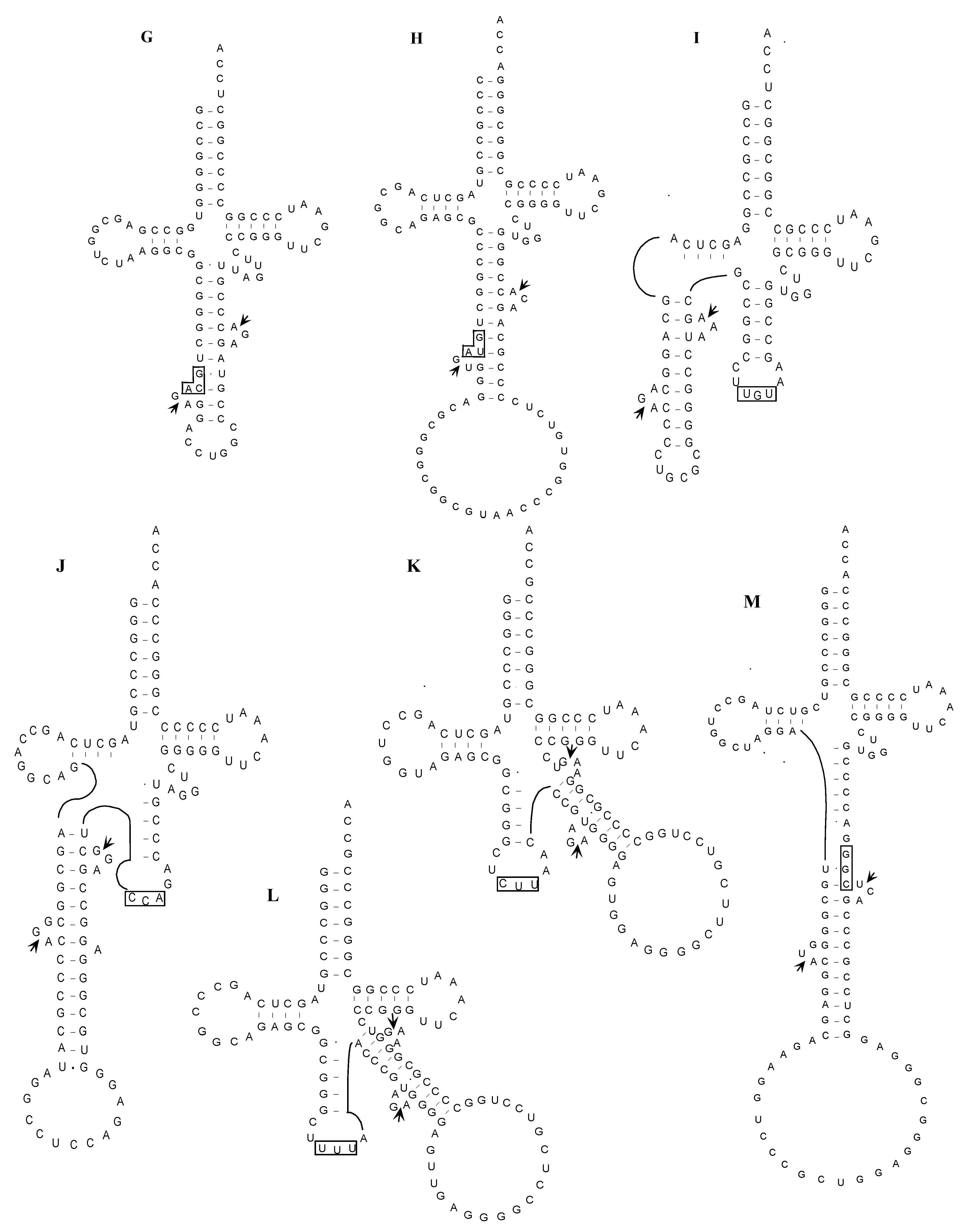
2.2. The Whole Set of tRNA Molecules in Sulfolobus tokodaii strain7
2.2.1. Introduction and Features of tRNAs in S. tokodaii strain7

| tRNA Species | un | sp | tRNA Species | un | sp | tRNA Species | un | sp | tRNA Species | un | sp |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phe(AAA) | - | - | Ser(AGA) | - | - | Tyr(AUA) | - | - | Cys(ACA) | - | - |
| Phe(GAA) | t | s | Ser(GGA) | - | HL | Tyr(GUA) | t | H | Cys(GCA) | t | s |
| Leu(UAA) | - | t | Ser(UGA) | t | HL | End(UUA) | - | - | End(UCA) | - | - |
| Leu(CAA) | t | t | Ser(CGA) | H | H | End(CUA) | - | - | Trp(CCA) | t | HL |
| Leu(AAG) | - | - | Pro(AGG) | - | - | His(AUG) | - | - | Arg(ACG) | - | - |
| Leu(GAG) | t | s | Pro(GGG) | t | HL | His(GUG) | - | t | Arg(GCG) | t | HL |
| Leu(UAG) | t | HL | Pro(UGG) | - | t | Gln(UUG) | - | t | Arg(UCG) | - | s |
| Leu(CAG) | t | HL | Pro(CGG) | t | t | Gln(CUG) | - | t | Arg(CCG) | - | t |
| Ile(AAU) | - | - | Thr(AGU) | - | - | Asn(AUU) | - | - | Ser(AGU) | - | - |
| Ile(GAU) | t | HL | Thr(GGU) | - | t | Asn(GUU) | - | t | Ser(GGU) | - | t |
| Ile(UAU) | - | - | Thr(UGU) | t | s | Lys(UUU) | HL | HL | Arg(UCU) | t | s |
| Met(CAU) # | t | s | Thr(CGU) | t | s | Lys(CUU) | t | s | Arg(CCU) | t | HL |
| Val(AAC) | - | - | Ala(AGC) | - | - | Asp(AUC) | - | - | Gly(ACC) | - | - |
| Val(GAC) | - | tL | Ala(GGC) | - | s | Asp(GUC) | - | t | Gly(GCC) | - | t |
| Val(UAC) | - | t | Ala(UGC) | - | s | Glu(UUC) | t | HL | Gly(UCC) | - | t |
| Val(CAC) | - | t | Ala(CGC) | - | t | Glu(CUC) | HL | HL | Gly(UCC) | - | tL |
2.2.2. PCR-Based Identification of tRNA Molecules
2.2.3. Identification of Cleavage Sites for Introns in tRNA Genes

3. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Andachi, Y.; Yamao, F.; Muto, A.; Osawa, S. Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. Resemblance to mitochondria. J. Mol. Biol. 1989, 209, 37–54. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R.; Fox, G.E. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef] [PubMed]
- Szekely, E.; Belford, H.G.; Greer, C.L. Intron sequence and structure requirements for tRNA splicing in Saccharomyces cerevisiae. J. Biol. Chem. 1988, 263, 13839–13847. [Google Scholar] [PubMed]
- Thompson, L.D.; Daniels, C.J. A tRNA(Trp) intron endonuclease from Halobacterium volcanii. Unique substrate recognition properties. J. Biol. Chem. 1988, 263, 17951–17959. [Google Scholar] [PubMed]
- Thompson, L.D.; Daniels, C.J. Recognition of exon-intron boundaries by the Halobacterium volcanii tRNA intron endonuclease. J. Biol. Chem. 1990, 265, 18104–18111. [Google Scholar] [PubMed]
- Sako, Y.; Nomura, N.; Uchida, A.; Ishida, Y.; Morii, H.; Koga, Y.; Hoaki, T.; Maruyama, T. Aeropurum pernix gen. nov., sp. nov., a novel aerobic hyper thermophilic archaeon growing at temperatures up to 100 °C. Int. J. Syst. Bacteriol. 1996, 46, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Kawarabayasi, Y.; Hino, Y.; Horikawa, H.; Yamazaki, S.; Haikawa, Y.; Jin-no, K.; Takahashi, M.; Sekine, M.; Baba, S.; Ankai, A.; et al. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeota, Aeropyrum pernix K1. DNA Res. 1999, 6, 84–76, 147–155. [Google Scholar]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, S.; Kikuchi, H.; Kawarabayasi, Y. Characterization of a whole set of tRNA molecules in an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 2006, 12, 403–416. [Google Scholar] [CrossRef]
- Chan, P.P.; Cozen, A.E.; Lowe, T.M. Discovery of permuted and recently split transfer RNAs in Archaea. Genome Biol. 2011, 12, R38. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Iwasaki, T.; Uzawa, T.; Hara, K.; Nemoto, N.; Kon, T.; Ueki, T.; Yamagishi, A.; Oshima, T. Sulfolobus tokodaii sp. nov. (f. Sulfolobus sp. strain7), a new member of the genus Sulfolobus isolated from Beppu Hot Springs, Japan. Extremophiles 2002, 6, 39–44. [Google Scholar]
- Kawarabayasi, Y.; Hino, Y.; Horikawa, H.; Jin-no, K.; Takahashi, M.; Sekine, M.; Baba, S.; Ankai, A.; Kosugi, H.; Hosoyama, A.; et al. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain7. DNA Res. 2001, 8, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Fichant, G.A.; Burks, C. Identifying potential tRNA genes in genomic DNA sequences. J. Mol. Biol. 1991, 220, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009, 37, D93–D97. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, S.; Yoshinari, S.; Kita, K.; Watanabe, Y.; Kawarabayasi, Y. Identification of an entire set of tRNA molecules and characterization of cleavage sites of the intron-containing tRNA precursors in acidothermophilic crenarchaeon Sulfolobus tokodaii strain7. Gene 2011, 489, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Hirata, A.; Kitajima, T.; Hori, H. Cleavage of intron from the standard or non-standard position of the precursor tRNA by the splicing endonuclease of Aeropyrum pernix, a hyper-thermophilic Crenarchaeon, involves a novel RNA recognition site in the Crenarchaea specific loop. Nucleic Acids Res. 2011, 39, 9376–9389. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Yoshinari, S. Intron and RNA splicing in Archaea. Viva Orig. 2013, 41, 12–13. [Google Scholar]
- Yoshinari, S.; Fujita, S.; Masui, R.; Kuramitsu, S.; Yokobori, S.; Kita, K.; Watanabe, Y. Functional reconstitution of a crenarchaeal splicing endonuclease in vitro. Biochem. Biophys. Res. Commun. 2005, 334, 1254–1259. [Google Scholar] [CrossRef] [PubMed]
- Marck, C.; Grosjean, H. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: Evolutionary implications. RNA 2003, 9, 1516–1531. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, J.; Kikuta, K.; Fujishima, K.; Yachie, N.; Tomita, M.; Kanai, A. Comprehensive analysis of archaeal tRNA genes reveals rapid increase of tRNA introns in the order thermoproteales. Mol. Biol. Evol. 2008, 25, 2709–2716. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watanabe, Y.-i.; Kawarabayasi, Y. Experimental Confirmation of a Whole Set of tRNA Molecules in Two Archaeal Species. Int. J. Mol. Sci. 2015, 16, 2187-2203. https://doi.org/10.3390/ijms16012187
Watanabe Y-i, Kawarabayasi Y. Experimental Confirmation of a Whole Set of tRNA Molecules in Two Archaeal Species. International Journal of Molecular Sciences. 2015; 16(1):2187-2203. https://doi.org/10.3390/ijms16012187
Chicago/Turabian StyleWatanabe, Yoh-ichi, and Yutaka Kawarabayasi. 2015. "Experimental Confirmation of a Whole Set of tRNA Molecules in Two Archaeal Species" International Journal of Molecular Sciences 16, no. 1: 2187-2203. https://doi.org/10.3390/ijms16012187





