2.1. Staphylococcus aureus-Induced Acute Rhinosinusitis in C57BL/6 Mice, and the Secretion of Interleukin (IL)-1β
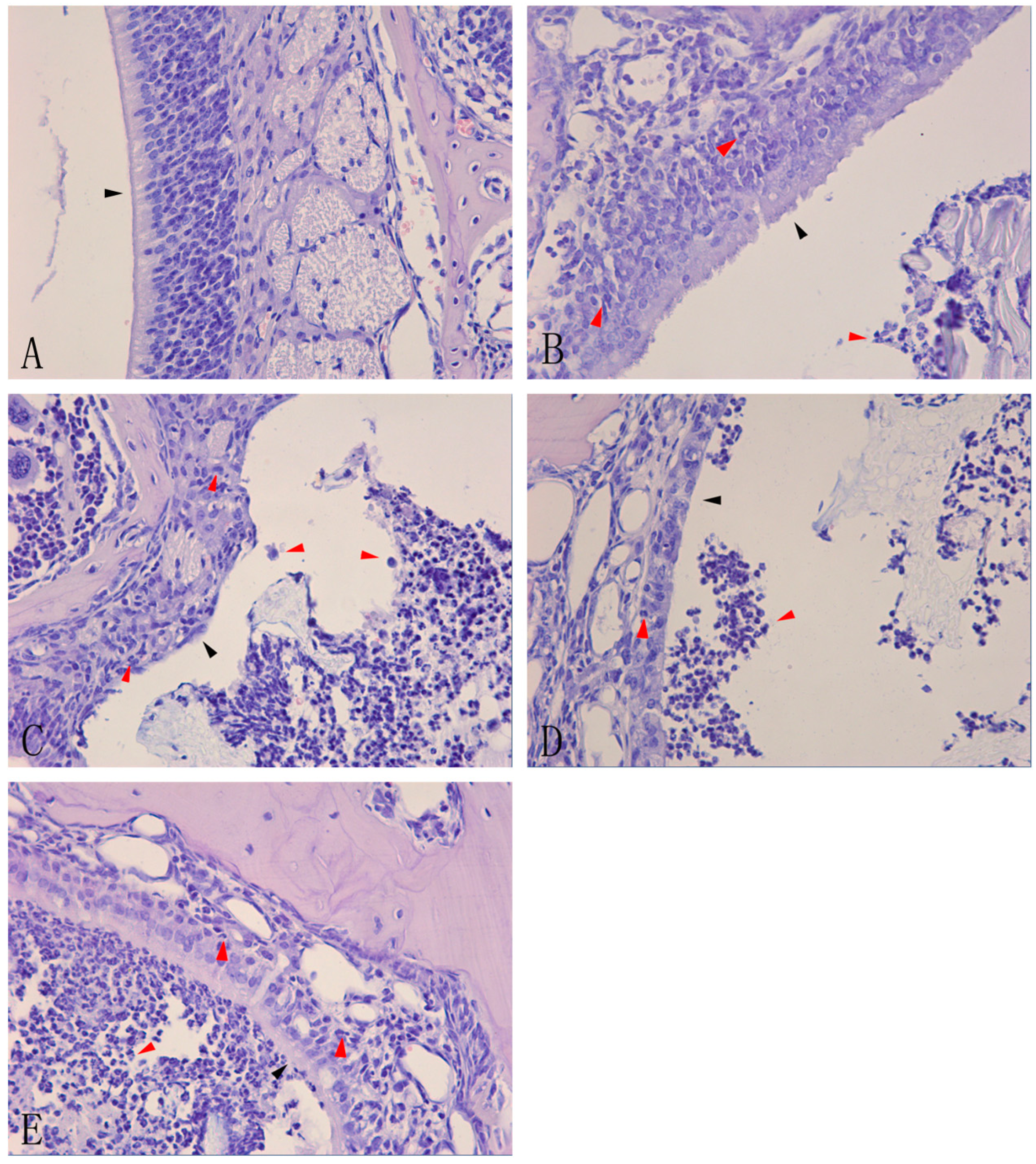
A previous study that attempted to establish a model of acute rhinosinusitis revealed that neutrophil clusters occupied the nasal sinus and that neutrophils infiltrated and damaged the nasal mucosa, as shown by hematoxylin-eosin staining [
13]. To test whether the model was successfully constructed, hematoxylin-eosin staining was carried out to determine the histological features of the nasal mucosa and sinus of mice.
A real-time polymerase chain reaction (PCR) was performed to measure the mRNA of the pro-inflammatory cytokine IL-1β, and western blot was used to detect the mature IL-1β protein in the nasal mucosa. Up-regulation of the expression level of IL-1β can be used to provide evidence of inflammation.
Histological examination of the nasal mucosa from the control group (group A,
n = 6) showed normal cells and a normal structure, and no inflammatory cell infiltration (
Figure 1A). However, gradually increasing infiltration of inflammatory cells and nasal damage to varying degrees were observed in groups B, C, D, and E (
n = 6 in each group). A slight infiltration of inflammatory cells and mild damage to the nasal mucosa were observed in group B, with a small number of neutrophils in the nasal sinus and cilia lodging of ciliated cells (
Figure 1B). The damage was more severe in group C, with more neutrophils in the nasal sinus, ciliated cell damage, and loss of cilia (
Figure 1C). In group D, there was a large accumulation of neutrophils within or around the nasal mucosa, loss of ciliated cells, and thinning of the mucosal layer (
Figure 1D). Group E had more neutrophils in the nasal sinus and nasal mucosa than group D, and the ciliated cells were severely destroyed (
Figure 1E).
Figure 1.
Hematoxylin-eosin staining of nasal mucosa of acute rhinosinusitis induced by Staphylococcus aureus.Controlmice remained un-inoculated (A). C57BL/6 mice were inoculated in the right nasal cavity with a suspension of S. aureus (10μL 1.2×109 CFU/mL) and the inflammation was analyzed after 1 day (B); 3days (C); 7 days (D); and 14 days (E).Histological analysis of leukocyte infiltration and morphological analysis of the nasal mucosa in the nasal cavity of mice were carried out by staining with hematoxylin-eosin (×40). The red arrows indicate infiltrated cells, and the black arrows indicate the damage to the nasal mucosa in images (B), (C), (D), and (E) and control nasal mucosa in image (A).
Figure 1.
Hematoxylin-eosin staining of nasal mucosa of acute rhinosinusitis induced by Staphylococcus aureus.Controlmice remained un-inoculated (A). C57BL/6 mice were inoculated in the right nasal cavity with a suspension of S. aureus (10μL 1.2×109 CFU/mL) and the inflammation was analyzed after 1 day (B); 3days (C); 7 days (D); and 14 days (E).Histological analysis of leukocyte infiltration and morphological analysis of the nasal mucosa in the nasal cavity of mice were carried out by staining with hematoxylin-eosin (×40). The red arrows indicate infiltrated cells, and the black arrows indicate the damage to the nasal mucosa in images (B), (C), (D), and (E) and control nasal mucosa in image (A).
Real-time PCR showed that IL-1β mRNA was rarely expressed in group A (
n = 6). One day after inoculation (group B,
n = 6), the expression of IL-1β mRNA was markedly increased compared with the control group (group A), and was statistically significant (
p < 0.05). Furthermore, the expression of IL-1β mRNA in groups B, C, D, and E (
n = 6 in each group) increased gradually and differed statistically significantly between groups A and B, groups D and E and groups A and E (
p < 0.05) (
Figure 2A). Western blot showed that the mature IL-1β protein was not expressed in the control group, but the level of expression of this protein in the nasal mucosa increased gradually from 1, 3, 7, and 14 days after stimulation, and differed statistically significantly in adjacent groups. (
p < 0.05) (
Figure 2B).
Figure 2.
The expression of IL-1βmRNA and mature IL-1β protein in the right nasal mucosa of mice. (A) IL-1β mRNA was rarely expressed in the control group; with time, the mRNA levels of IL-1βin the nasal mucosa after 1, 3, 7, and 14 days following stimulationwith S. aureus gradually increased;and (B) Mature IL-1β protein was not expressed in the control group; after inoculation, the protein levels of mature IL-1βin the nasal mucosa after 1, 3, 7, and 14 days following stimulationwith S. aureus gradually increased (*indicates p < 0.05; ns= not statistically significant).
Figure 2.
The expression of IL-1βmRNA and mature IL-1β protein in the right nasal mucosa of mice. (A) IL-1β mRNA was rarely expressed in the control group; with time, the mRNA levels of IL-1βin the nasal mucosa after 1, 3, 7, and 14 days following stimulationwith S. aureus gradually increased;and (B) Mature IL-1β protein was not expressed in the control group; after inoculation, the protein levels of mature IL-1βin the nasal mucosa after 1, 3, 7, and 14 days following stimulationwith S. aureus gradually increased (*indicates p < 0.05; ns= not statistically significant).
2.2 NLR Pyrin Domain Containing 3 (NLRP3) Increased with Time
The expression of NLRP3 increased to varying degrees with time. At the protein level, western blot showed that the expression of NLRP3 protein in groups A, B, C, D, and E (
n = 6 in each group) gradually increased and differed statistically significantly (
p < 0.05) between groups E and D, and groups E and A, (
Figure 3A). Immunofluorescence displayed a similar trend (
Figure 3B). Real-time PCR showed that expression of NLRP3 mRNA in groups A, B, C, D, and E increased gradually and statistically significantly in adjacent groups, except for groups C and B (
p < 0.05) (
Figure 3C).
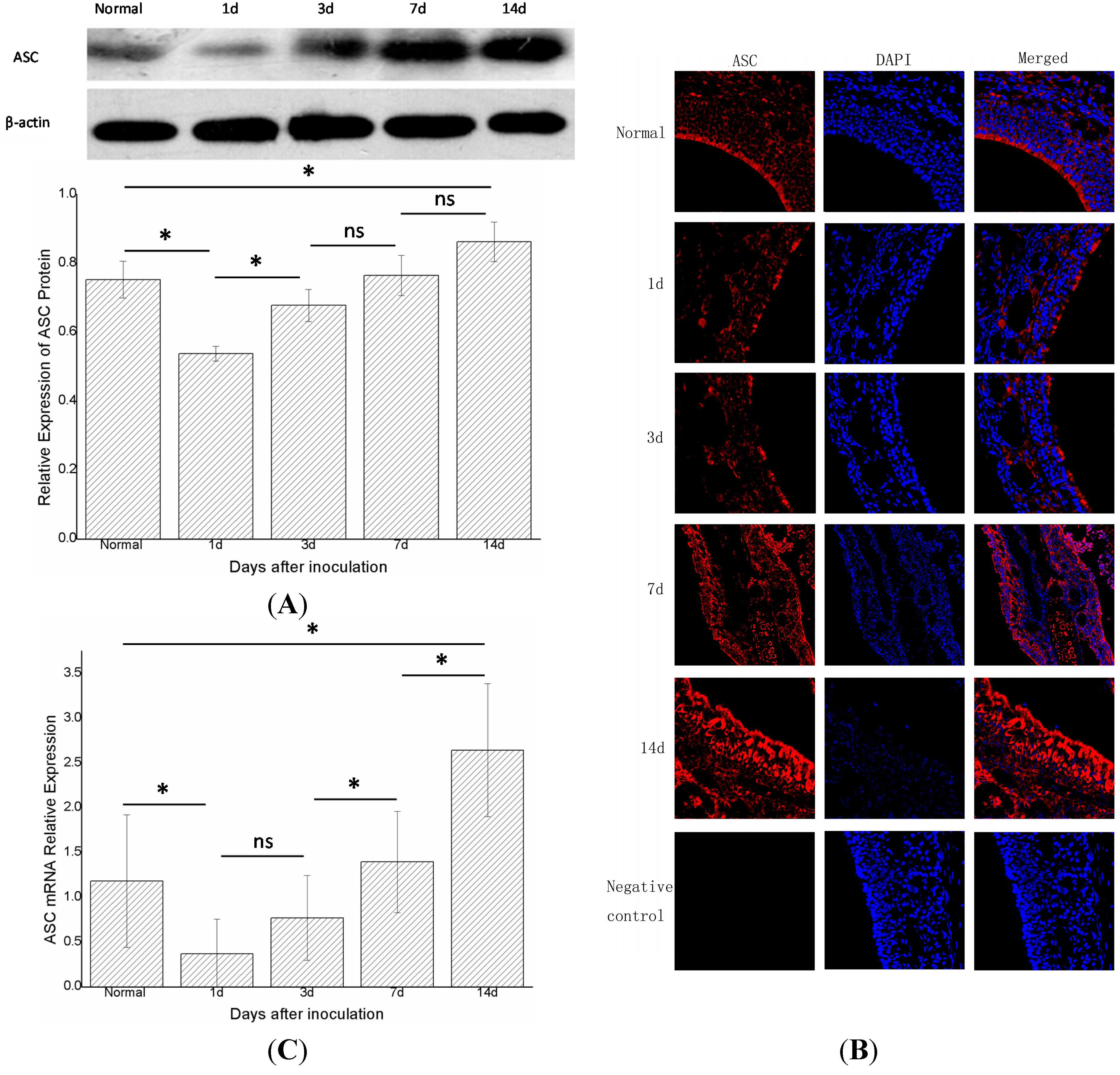
2.3. Apoptosis-Associated Speck-Like Protein (ASC) Decreased at First and then Increased again
The changes in ASC did not parallel those of NLRP3. One day after inoculation (group B,
n = 6), the expression of the ASC protein decreased compared with the control group (group A,
n = 6) (
p < 0.05). In groups B, C, D, and E (
n = 6 in each group), the expression of ASC protein increased gradually with time. Moreover, the levels of ASC protein differed statistically significantly (
p < 0.05) between groups C and B, groups E and A (
Figure 4A). Immunofluorescence showed a similar trend (
Figure 4B). Real-time PCR showed that the expression of ASC mRNA in group B declined compared with the control group (group A,
n = 6) (
p < 0.05). In groups B, C, D, and E (
n = 6 in each group), the expression gradually increased with time, and differed statistically significantly in adjacent groups, except for groups B and C (
p < 0.05) (
Figure 4C).
Figure 3.
The expression of total NLRP3 protein and NLRP3 mRNA in the right nasal mucosa of mice.(A) Western blot assessment of the protein expression of NLRP3 in the control group (group A), and 1, 3, 7, and 14 days (groups B, C, D, and E, respectively) after stimulation with S. aureus. A semi-quantitative analysis was used to represent the total protein level of NLRP3; (B) Immunofluorescence showed that NLRP3was mainly expressed in the cytoplasm of cells in the nasal mucosa; and (C) Real-time PCR assessment of the mRNA expression of NLRP3 in the control group(group A), and 1,3,7, and 14days (groups B, C, D, and E, respectively) after stimulationwith S. aureus (* indicates p < 0.05; ns= not statistically significant).
Figure 3.
The expression of total NLRP3 protein and NLRP3 mRNA in the right nasal mucosa of mice.(A) Western blot assessment of the protein expression of NLRP3 in the control group (group A), and 1, 3, 7, and 14 days (groups B, C, D, and E, respectively) after stimulation with S. aureus. A semi-quantitative analysis was used to represent the total protein level of NLRP3; (B) Immunofluorescence showed that NLRP3was mainly expressed in the cytoplasm of cells in the nasal mucosa; and (C) Real-time PCR assessment of the mRNA expression of NLRP3 in the control group(group A), and 1,3,7, and 14days (groups B, C, D, and E, respectively) after stimulationwith S. aureus (* indicates p < 0.05; ns= not statistically significant).
Figure 4.
The expression of total ASC protein and ASC mRNA in the right nasal mucosa of mice. (A) Western blot assessment of the protein expression of ASC in the control group (group A),and1,3,7,and 14 days (groups B, C, D, and E, respectively) after stimulationwith S. aureus. A semi-quantitative analysis was used to represent the total protein level of ASC; (B) Immunofluorescence showed that ASC wasmainly expressed in the cytoplasm of cells in the nasal mucosa;and (C) Real-time PCR assessment of the mRNA expression of ASC in the control group (group A), and 1,3,7,and 14 days (groups B, C, D, and E, respectively) after stimulationwith S. aureus (* indicates p < 0.05; ns= not statistically significant).
Figure 4.
The expression of total ASC protein and ASC mRNA in the right nasal mucosa of mice. (A) Western blot assessment of the protein expression of ASC in the control group (group A),and1,3,7,and 14 days (groups B, C, D, and E, respectively) after stimulationwith S. aureus. A semi-quantitative analysis was used to represent the total protein level of ASC; (B) Immunofluorescence showed that ASC wasmainly expressed in the cytoplasm of cells in the nasal mucosa;and (C) Real-time PCR assessment of the mRNA expression of ASC in the control group (group A), and 1,3,7,and 14 days (groups B, C, D, and E, respectively) after stimulationwith S. aureus (* indicates p < 0.05; ns= not statistically significant).
2.4. Caspase-1 mRNA Decreased, then Increased; the Protein of the Fragment of Procaspase-1 (p20) Increased over Time
After the NLRP3 inflammasome was activated, procaspase-1 was activated into the form of active caspase-1, which is a tetramer composed of p20 and p10. Therefore, the detection of p20 and p10 can reflect whether the NLRP3 inflammasome has been activated. Western blot showed that procaspase-1 decreased in groups B and C compared with the control group (group A,
n = 6) and then later increased in group D (
p < 0.05). In groups B, C, D, and E (
n = 6 in each group), the expression of procaspase-1 protein was increased and differed significantly in adjacent groups, except for groups E and D (
p < 0.05) (
Figure 5A). Immunofluorescence showed a similar trend (
Figure 5B). In this experiment, we measured p20 protein to determine whether the NLRP3 inflammasome was activated. Western blot showed that the p20 protein was not expressed in the control group (group A), but increased gradually in groups B, C, D, and E, and differed statistically significantly between the adjacent groups (
p < 0.05). Real-time PCR showed that the expression of caspase-1 mRNA in group B was lower than that in the control group (group A,
n = 6). In groups B, C, D, and E (
n = 6 in each group), the expression of caspase-1 mRNA gradually increased and differed statistically significantly between groups E and D , and groups E and A. (
p < 0.05) (
Figure 5C).
2.5. Discussion
In this study, we constructed a mouse model of acute rhinosinusitis. Hematoxylin-eosin staining revealed ongoing infiltration of inflammatory cells and damage to the nasal mucosa (
Figure 1). This result was in accord with previous studies that suggested that the inflammatory cells represented acute rhinosinusitis [
13,
14]. The gradually increased expression level of IL-1β mRNA and mature IL-1β protein provided more powerful proof that the construction of this mouse model of acute rhinosinusitis was successful.
The mechanisms of acute rhinosinusitis are not clear. In this study, we focused on the innate immune system for it may involve in the process of acute rhinosinusitis, and the innate immune system is generally considered to function via PRRs. To date, the most widely studied PRRs have been the TLRs, such as TLR-2, TLR-4, and TLR-9 [
4,
15]. However, another type of PRR, called NLR, was related to rhinosinusitis and was found in the cytoplasm. NLRP3 was the most important member of the NLRs [
16], and was integrated with ASC through a PYD (pyrin domain)–PYD interaction. The ASC then combined with caspase-1 through a CARD (caspase recruitment domain–CARD interaction. At this time, the caspase-1was not yet activated, and was also known as procaspase-1. The combined NLRP3-ASC-procaspase-1 complex was termed the NLRP3 inflammasome. The combined procaspase-1 was activated to release splitting fragments p20 and p10, which formed a tetramer that was the active caspase-1. The active caspase-1 assisted IL-1β and IL-18 to convert to their mature forms, which were then secreted with the active caspase-1. Thus, p20 expression can reflect whether procaspase-1 is activated. However, in the state of inflammation in mice, not only the NLRP3 inflammasome can active procaspsae-1; there are other pathways, such as the AIM2 (absent in melanoma 2) inflammasome pathway. Therefore, we also investigated NLRP3 and ASC to determine whether the NLRP3 inflammasome participates in the signal transduction. Thus, we measured NLRP3, ASC, procaspase-1 and p20 to determine the expression levels of the assembled NLRP3 inflammasome [
17]. Moreover, we investigated the presence of IL-1β mRNA and mature IL-1β protein to verify whether the active caspase-1 is generated. The NLRP3 inflammasome was indicated to play a pro-inflammatory role in this model after inoculation with bacteria; the level of NLRP3 protein increased constantly after inoculation with
S. aureus (
Figure 3A), and the other two components of the NLRP3 inflammasom––ASC protein (
Figure 4A) and procaspase-1 protein (
Figure 5A) showed the same trend. The results of immunofluorescence were consistent with those of the western blot (
Figure 3B,
Figure 4B and
Figure 5B). These results revealed that the NLRP3 inflammasome was activated after inoculation, and that the expression level of the assembled NLRP3 inflamasome in groups B, C, D and E increased gradually. It could therefore be concluded that the NLRP3 inflammasome was activated in this model, and there was a positive correlation between the expression level of the NLRP3 inflammasome and the severity of acute rhinosinusitis.
Real-time PCR found that the mRNAs of NLRP3, ASC, and caspase-1 gradually increased after infection (
Figure 3C,
Figure 4C and
Figure 5C), as did the mRNA of IL-1β (
Figure 2A). A previous study suggested that active caspase-1 was an IL-1β converting enzyme, and directly promoted the maturation of IL-1β [
18]. Moreover, IL-1β is a common pro-inflammatory cytokine that is deemed to be an index of the severity of inflammation. Thus, we considered that the NLRP3 inflammasome was activated in this model and its expression level was positively correlated with the severity of acute rhinosinusitis at the gene level.
Figure 5.
The expression of caspase-1 protein (also called procaspase-1 before caspase-1 was activated) and the fragment of procaspase-1 (p20) and caspase-1 mRNA in the right nasal mucosa of mice. (A) Western blot assessment of the protein expression of procaspase-1 and p20 in the control group (group A), and1, 3, 7, and 14 days (groups B,C,D and E, respectively) after stimulation with Staphylococcus aureus. A semi-quantitative analysis was used to represent the total protein level ofprocaspase-1 and p20; (B) Immunofluorescence showed that caspase-1 was mainly expressed in the cytoplasm of cells in the nasal mucosa; and (C) Real-time PCR assessment of the mRNA expression of caspase-1 in the control group (group A), and 1, 3, 7, and 14 days (groups B, C, D, and E, respectively) after stimulationwith Staphylococcus aureus (* indicates p < 0.05; ns= not statistically significant).
Figure 5.
The expression of caspase-1 protein (also called procaspase-1 before caspase-1 was activated) and the fragment of procaspase-1 (p20) and caspase-1 mRNA in the right nasal mucosa of mice. (A) Western blot assessment of the protein expression of procaspase-1 and p20 in the control group (group A), and1, 3, 7, and 14 days (groups B,C,D and E, respectively) after stimulation with Staphylococcus aureus. A semi-quantitative analysis was used to represent the total protein level ofprocaspase-1 and p20; (B) Immunofluorescence showed that caspase-1 was mainly expressed in the cytoplasm of cells in the nasal mucosa; and (C) Real-time PCR assessment of the mRNA expression of caspase-1 in the control group (group A), and 1, 3, 7, and 14 days (groups B, C, D, and E, respectively) after stimulationwith Staphylococcus aureus (* indicates p < 0.05; ns= not statistically significant).
The activation of the NLRP3 inflammasome could be induced by many factors, such as pore-forming toxins and α-hemolysin produced by
S. aureus [
19,
20]. Once these agonists enabled the NLRP3 inflammasome, caspase-1 was activated and induced the maturation and release of IL-1β and IL-18, resulting in inflammation. However, the mechanism was considered to be controversial. Three hypotheses on the mechanism of activation of the NLRP3 inflammasome have recently been proposed. The first hypothesis was that the P2X7 ion channel was activated, potassium ions leaked out, resulting in cell membrane perforation, and then the agonists entered the cytosol to activate the NLRP3 inflammasome. The second hypothesis suggested that the lysosome was activated by the agonists and released cathepsin B, which combined with NLRP3 to activate the NLRP3 inflammasome. The third hypothesis proposed that the agonists caused the formation of reactive oxygen species, which activated the NLRP3 inflammasome [
17]. In our study, we observed that the NLRP3 inflammasome was activated and a continual increase in the NLRP3 inflammasome after inoculation with bacteria. We presumed that the NLRP3 inflammasome was activated resulting in an inflammatory cascade after inoculation, and then some endogenous NLRP3 agonists, for example, adenosine triphosphate and glucose, were released and activated more NLRP3 inflammasomes [
19,
21]. In addition, our previous study found that the probability of the formation of biofilm was increased after the mice were inoculated, and thus the pathogenicity of
S. aureus could be enhanced due to the protection of the biofilm [
22].
An interesting phenomenon was that ASC and procaspase-1 had reduced protein and mRNA levels one day after inoculation (
Figure 4 and
Figure 5) compared with the control group (no bacterial inoculation). Similar results showing a decrease in ASC levels after cytokine induction by
Porphyromonas gingivalis-infected human THP1 monocytic cells were reported by Taxman
et al. [
23]. We speculated that some substances or some pathways depressed the levels of ASC and procaspase-1. A previous study found that pathogens activated inducible nitric oxide synthase (iNOS) to produce nitric oxide (NO) in the macrophages of mice, which could depress the activation of the NLRP3 inflammasome by preventing ASC pyroptosome formation and inhibiting the activation of caspase-1 [
24]. Moreover, it has been reported that
S. aureus could induce the production of NO by activating the iNOS [
25]. We therefore inferred that NO was produced after the activation of iNOS in our experiment, resulting in the prevention of ASC pyroptosome formation and the inhibition of caspase-1 activation. In addition, procaspase-1 could be directly inhibited by superoxide [
26]. This type of material could also explain the decrease in procaspase-1 after inoculation compared with controls. The later increases in the expression of ASC and procaspase-1 might be the outcome of the inhibiting effects of these two components could be diminished by decreasing of NO, while decreasing of NO maybe due to increased adenosine triphosphate depleted iNOS [
24,
27]. The pro-inflammatory effects enhanced by increased adenosine triphosphate and bacterial biofilm formation also could have contributed to the later increases in the expression of ASC and procaspase-1. However, IL-1β mRNA increased at the same time, possibly because not only the NLRP3 inflammasome but also another factor enhanced the expression of IL-1β, such as proteinase-3 and elastase [
28]. The exact explanation still needs to be explored in further studies.