New Milk Protein-Derived Peptides with Potential Antimicrobial Activity: An Approach Based on Bioinformatic Studies
Abstract
:1. Introduction
2. Results and Discussion
2.1. Physico-Chemical Characteristic of AMPs
| Amino Acid | AMPs in BIOPEP | AMPs from Milk Proteins | ||||||
|---|---|---|---|---|---|---|---|---|
| Average Amino Acid Content (%) | Number of Peptides Containing Given Amino Acid | Min.-Max. Amino Acid Content (%) | Average Amino Acid Content (%) | Number of Peptides Containing Given Amino Acid | Min.-Max Amino Acid Content (%) | |||
| a | b | a | b | |||||
| Ala | 6.8 | 9.8 | 286 | 0–33.3 | 6.4 | 10.6 | 36 | 0–25.0 |
| Arg | 8.2 | 12.3 | 278 | 0–33.3 | 7.0 | 11.9 | 35 | 0–25.0 |
| Asn | 3.4 | 6.1 | 234 | 0–25.0 | 2.0 | 9.7 | 12 | 0–25.0 |
| Asp | 2.3 | 5.7 | 166 | 0–25.0 | 2.6 | 14.0 | 11 | 0–25.0 |
| Cys | 5.8 | 13.6 | 176 | 0–40.0 | 3.0 | 10.4 | 17 | 0–25.0 |
| Gln | 3.7 | 7.1 | 220 | 0–25.0 | 7.0 | 11.4 | 36 | 0–33.3 |
| Glu | 2.3 | 5.6 | 175 | 0–25.0 | 4.4 | 11.3 | 23 | 0–25.0 |
| Gly | 10.5 | 12.5 | 349 | 0–63.1 | 3.3 | 9.3 | 21 | 0–20.0 |
| His | 2.1 | 5.8 | 152 | 0–18.4 | 1.2 | 6.1 | 12 | 0–14.3 |
| Ile | 6.1 | 8.1 | 312 | 0–40.0 | 6.5 | 10.2 | 38 | 0–33.3 |
| Leu | 9.9 | 11.4 | 339 | 0–58.3 | 8.7 | 12.3 | 42 | 0–37.5 |
| Lys | 10.3 | 12.7 | 337 | 0–62.5 | 10.2 | 15.5 | 19 | 0–33.3 |
| Met | 1.0 | 3.7 | 113 | 0–7.4 | 1.0 | 3.6 | 16 | 0–7.1 |
| Phe | 4.0 | 6.2 | 258 | 0–37.5 | 2.6 | 7.3 | 21 | 0–16.7 |
| Pro | 5.0 | 9.7 | 215 | 0–53.2 | 8.0 | 11.8 | 40 | 0–28.6 |
| Ser | 5.1 | 7.5 | 282 | 0–23.1 | 4.0 | 9.1 | 26 | 0–21.4 |
| Thr | 3.4 | 6.3 | 226 | 0–33.3 | 5.3 | 10.8 | 29 | 0–33.3 |
| Trp | 2.1 | 5.0 | 178 | 0–38.5 | 3.2 | 8.0 | 24 | 0–16.7 |
| Tyr | 2.6 | 7.0 | 157 | 0–33.3 | 5.6 | 14.4 | 23 | 0–33.3 |
| Val | 5.9 | 8.2 | 300 | 0–37.3 | 8.0 | 12.5 | 38 | 0–37.5 |
| Index | AMPs Collected in BIOPEP | AMPs from Milk Proteins | ||||
|---|---|---|---|---|---|---|
| Mean Value | Min.–Max. Value | Predominant Value (%) | Mean Value | Min.–Max. Value | Predominant Value (%) | |
| Molecular mass (Da) | 3242.9 | 393.5–14,350.8 | 2000–4000 (47) | 1906.9 | 393.5–6707.4 | 393–1000 (39) |
| pI | 9.3 | 3.4–13.3 | 9–10 (29) | 8.1 | 3.4–12.0 | 10–11 (25) |
| Net charge | 3.7 | −7.0–20 | 0–5 (56) | 2.2 | −7.0–10.1 | −2–0 (34) |
| Instability index | 31.7 | −50.4–166.2 | 0–60 (71) | 41.5 | −30.9–157.7 | 0–20 (24) |
| Aliphatic index | 83.9 | 0–227.5 | 40–120 (65) | 89.6 | 0.0–226.7 | 60–100 (49) |
| GRAVY | −0.2 | −3.51–3.6 | −1–0 (47) | −0.4 | −2.5–2.2 | −1–0 (49) |
| Boman Index (kcal/mol) | 1.5 | −2.6–6.8 | 1–2 (28) | 1.3 | −6.9–5.0 | 1–3 (61) |
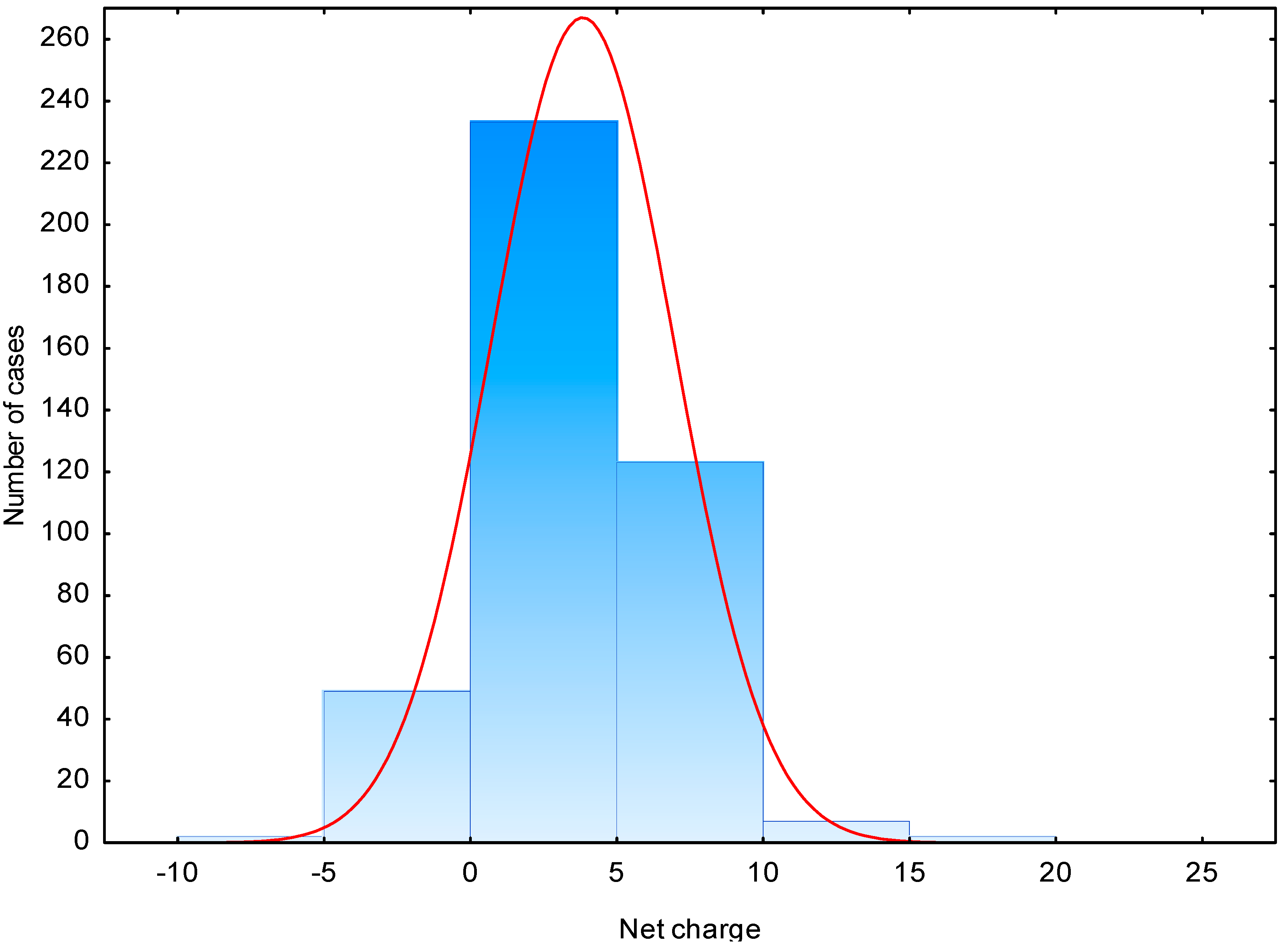
2.2. In Silico Proteolysis of Milk Proteins
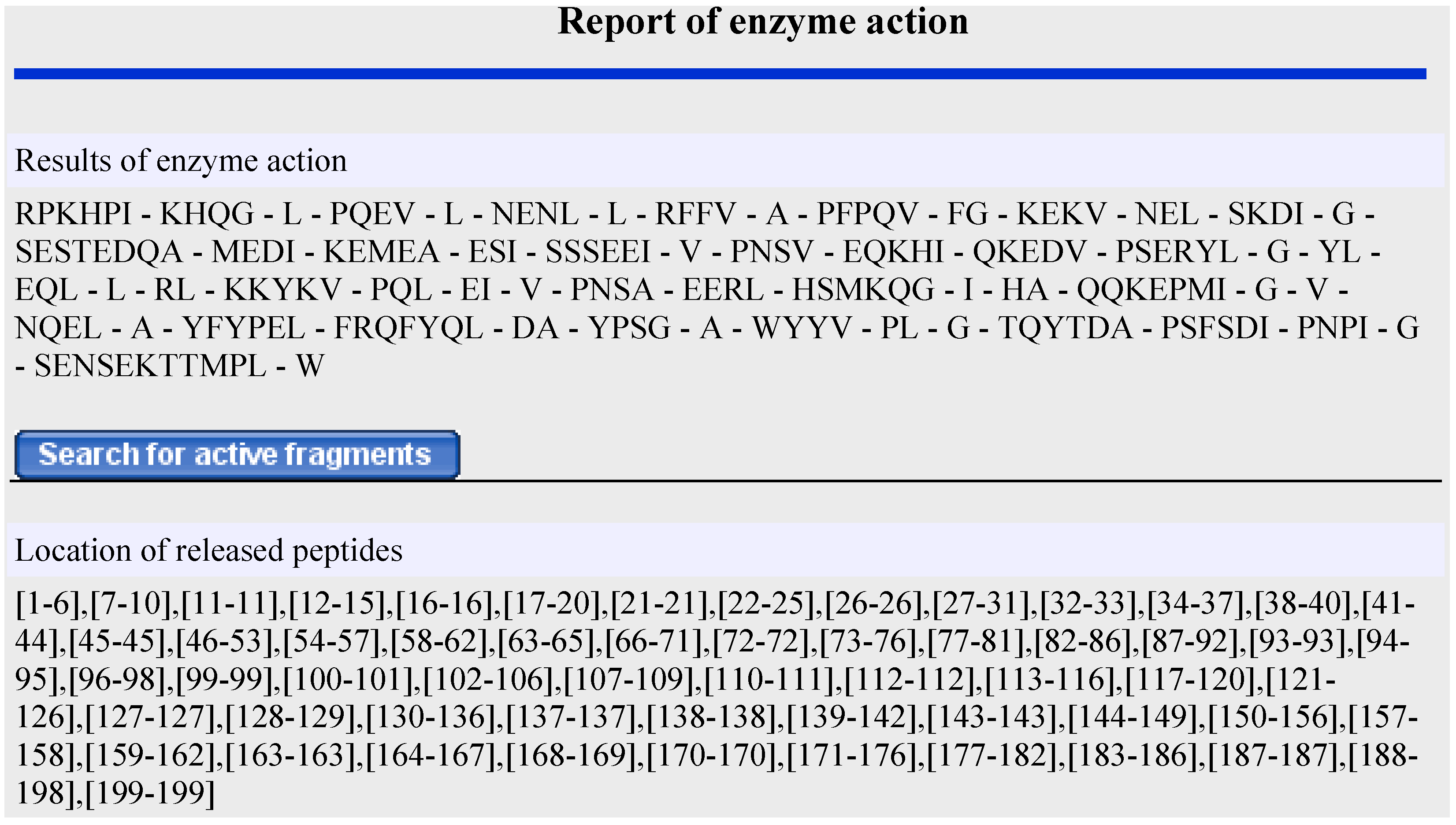
2.3. Prediction of Antimicrobial Activity of Peptides Released During in Silico Proteolysis of Milk Proteins
| Sequence | AMP Origin/Position | Net Charge | Isoelectric Point pH | Molecular Mass (Da) | Boman Index | Instability Index | Aliphatic Index | GRAVY | SVM a | RFC b | ANN c | DAC d |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DDKHYQKA (Pancreatic elastase EC 3.4.21.36, Leukocyte elastase EC 3.4.21.37) | αs2-casein, gen. var. A-11P f(74–81) | 0.1 | 7.72 | 1004.07 | 4.63 | 53.06 | 12.50 | −2.625 | 1.000 | 0.622 | NAMP | 0.507 |
| GQRDLLFKDSALGFLRIP (Prolyl oligopeptidase EC 3.4.21.26) | Lactoferrin f(294–311) | 1.0 | 10.08 | 2046.41 | 1.52 | 20.89 | 113.89 | 0.028 | 0.659 | 0.4835 | AMP | 0.834 |
| ADALNLDGGYIYTAGKCGLVPVLAE (V-8 protease EC 3.4.21.19) | Lactoferrin f(389–413) | −2.0 | 3.7 | 2523.89 | −0.22 | 16.36 | 117.20 | 0.536 | 0.568 | 0.5215 | NAMP | 0.816 |
| QEQNQEQP (Prolyl oligopeptidase EC 3.4.21.26), Thermolysin EC 3.4.24.27) | κ-casein, gen. var. A f(1–8) | −2.0 | 3.79 | 999.9 | 5.3 | 119.70 | 0.00 | −3.263 | 0.976 | 0.5845 | AMP | 0.000 |
| KKYKVPQL (Pepsin 1.3 EC 3.4.23.1, Pancreatic elastase EC 3.4.21.71) | αs1-casein, gen. var. B-8P f(102–109) | 3.0 | 10.45 | 1003.25 | 1.67 | 46.29 | 85.00 | −1.262 | 0.952 | 0.509 | NAMP | 0.916 |
| AVAVVKKGSNF (Chymase EC 3.4.212.39, Metridin EC 3.4.21.3) | Lactoferrin f(94–104) | 2.0 | 10.6 | 1119.33 | 0.13 | −14.91 | 97.27 | 0.591 | 0.943 | 0.483 | AMP | 0.912 |
| EMPFPK(Ficain EC 3.4.22.3, Bromelain EC 3.4.22.4) | β-casein, gen. var. A2-5P f(108–113) | 0.0 | 6.94 | 747.91 | 1.17 | 145.77 | 0.00 | −0.983 | 1.000 | 0.571 | AMP | 0.190 |
| EPEQSL (Ficain EC 3.4.22.3) | β-lactoglobulin gen. var. B f(112–117) | −2.0 | 3.13 | 701.73 | 2.94 | 174.73 | 65.00 | −1.517 | 1.000 | 0.4695 | NAMP | 0.569 |
| ITRINKKIEKFQS (Leukocyte elastase EC 3.4.21.37) | β-casein, gen. var. A2-5P f(23–35) | 3.0 | 10.83 | 1604.92 | 2.98 | 88.52 | 90.00 | −0.915 | 0.492 | 0.45 | AMP | 0.739 |
| ITRINKKIEKF (Proteinase P1 (lactocepin) EC 3.4.21.96) | β-casein, gen. var. A2-5P f(23–33) | 3.0 | 10.83 | 1389.71 | 2.71 | 62.85 | 106.36 | −0.691 | 0.694 | 0.46 | AMP | 0.879 |
| ALFGKNGKNCPDKFCLFK (Proteinase P1 (lactocepin) EC 3.4.21.96) | Lactoferrin f(616–633) | 2.9 | 9.71 | 2030.45 | 1.06 | −5.79 | 48.89 | −0.317 | 0.706 | 0.7655 | AMP | 0.987 |
3. Experimental Section
3.1. Materials
3.2. Physicochemical Properties of AMPs
3.3. In Silico Proteolysis of Milk Proteins
3.4. Prediction of Antimicrobial Activity of Peptides Released during in Silico Proteolysis of Milk Proteins
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Floris, R.; Recio, I.; Berkhout, B.; Visser, S. Antibacterial and antiviral effects of milk proteins and derivatives thereof. Curr. Pharm. Des. 2003, 9, 1257–1275. [Google Scholar] [CrossRef]
- Keymanesh, K.; Soltani, S.; Sardari, S. Application of antimicrobial peptides in agriculture and food industry. World J. Microbiol. Biotechnol. 2009, 25, 933–944. [Google Scholar] [CrossRef]
- Clare, D.A.; Swaisgood, H.E. Bioactive milk peptides: A prospectus. J. Dairy Sci. 2000, 83, 1187–1195. [Google Scholar] [CrossRef]
- Mils, S.; Ross, R.P.; Hill, C.; Fitzgerald, G.F.; Stanton, C. Milk intelligence: Mining milk for bioactive substances associated with human health. Int. Dairy J. 2011, 21, 377–401. [Google Scholar] [CrossRef]
- Dziuba, B. Antimicrobial peptides. In Bioactive Peptides and Food Proteins; Dziuba, J., Fornal, Ł., Eds.; WNT: Warsaw, Poland, 2009; pp. 35–46. [Google Scholar]
- Giuliani, A.; Pirri, G.; Nicoletto, S.F. Antimicrobial peptides: An overview of a promising class of therapeutics. Cent. Eur. J. Biol. 2007, 2, 1–33. [Google Scholar] [CrossRef]
- Li-Chan, E.C.Y. Antimicrobial peptides. In Nutraceutical Proteins and Peptides in Health and Disease; Mine, Y., Shahidi, F., Eds.; Taylor & Francis Group, CRC: New York, NY, USA, 2006; pp. 99–136. [Google Scholar]
- Wang, Z.; Wang, G. APD: The antimicrobial peptide database. Nucleic Acids Res. 2004, 32, D590–D592. [Google Scholar] [CrossRef]
- Hilpert, K.; Fjell, C.D.; Cherkasov, A. Short linear cationic antimicrobial peptides: Screening, optimizing, and prediction. In Peptide-Based Drug Design; Humana Press: Totowa, NJ, USA, 2008; pp. 127–159. [Google Scholar]
- Pellegrini, A. Antimicrobial peptides from food proteins. Curr. Pharm. Des. 2003, 9, 1225–1238. [Google Scholar] [CrossRef]
- Dashper, S.G.; Liu, S.W.; Reynolds, E.C. Antimicrobial peptides and their potential as oral therapeutic agents. Int. J. Pept. Res. Ther. 2007, 13, 505–516. [Google Scholar] [CrossRef]
- Hayes, M.; Ross, R.P.; Fitzgerald, G.F.; Hill, C.; Stanton, C. Casein-derived antimicrobial peptides generated by Lactobacillus acidophilus DPC6026. Appl. Environ. Microbiol. 2006, 72, 2260–2264. [Google Scholar] [CrossRef]
- López-Expósito, I.; Recio, I. Antibacterial activity of peptides and folding variants from milk proteins. Int. Dairy J. 2006, 16, 1294–1305. [Google Scholar] [CrossRef]
- Minervini, F.; Algaron, F.; Rizzello, C.G.; Fox, P.F.; Monnet, V.; Gobbetti, M. Angiotensin I-converting-enzyme-inhibitory and antibacterial peptides from Lactobacillus helveticus PR4 proteinase-hydrolyzed caseins of milk from six species. Appl. Environ. Microbiol. 2003, 69, 5297–5305. [Google Scholar] [CrossRef]
- Recio, I.; Visser, S. Identification of two distinct antibacterial domains within the sequence of bovine αs2-casein. Biochim. Biophys. Acta 1999, 1428, 314–326. [Google Scholar] [CrossRef]
- Tomita, M.; Wakabayashi, H.; Yamauchi, K.; Teraguchi, S.; Hayasawa, H. Bovine lactoferrin and lactoferricin derived from milk: Production and applications. Biochem. Cell Biol. 2002, 80, 109–112. [Google Scholar] [CrossRef]
- López-Expósito, I.; Quirós, A.; Amigo, L.; Recio, I. Casein hydrolysates as a source of antimicrobial, antioxidant and antihypertensive peptides. Lait 2007, 87, 241–249. [Google Scholar] [CrossRef]
- Dziuba, M.; Dziuba, B. In silico analysis of bioactive peptides. In Bioactive Proteins and Peptides as Functional Foods and Nutraceuticals; Mine, Y., Li-Chan, E.C.Y., Jiang, B., Eds.; Blackwell Publishing Ltd. and Institute of Food Technologists: London, UK, 2010; pp. 325–340. [Google Scholar]
- Thomas, S.; Karnik, S.; Barai, R.S.; Jayaraman, V.K.; Idicula-Thomas, S. CAMP: A useful resource for research on antimicrobial peptides. Nucleic Acids Res. 2010, 38, D774–D780. [Google Scholar] [CrossRef]
- BIOPEP. Available online: http://www.uwm.edu.pl/biochemia/index.php/pl/biopep (accessed from 2 to 31 January 2014).
- Epand, R.M.; Vogel, H.J. Diversity of antimicrobial peptides and their mechanisms of action. BBA Biomembr. 1999, 1462, 11–28. [Google Scholar]
- Laverty, G.; Gorman, S.P.; Gilmore, B.F. The Potential of antimicrobial peptides as biocides. Int. J. Mol. Sci. 2011, 12, 6566–6596. [Google Scholar] [CrossRef] [Green Version]
- Zamiatnin, A.A.; Poronina, O.L. Computer biochemistry and molecular physiology of the natural peptide ligands: different functional groups. In Peptide Science—Present and Future. Proceedings of the 1st International Peptide Symposium; Shimonishi, Y., Ed.; Kluwer: Boston, MA, USA, 1999; pp. 212–214. [Google Scholar]
- Akalin, A.S. Dairy-derived antimicrobial peptides: Action mechanisms, pharmaceutical uses and production proposals. Trends Food Sci. Technol. 2014, 36, 79–95. [Google Scholar] [CrossRef]
- Clare, D.A.; Catignani, G.L.; Swaisgood, H.E. Biodefense properties of milk: The role of antimicrobial proteins and peptides. Curr. Pharm. Des. 2003, 9, 1239–1255. [Google Scholar] [CrossRef]
- Lahov, E.; Regelson, W. Antibacterial and immunostimulating casein-derived substances from milk: Casecidin, isracidin peptides. Food Chem. Toxicol. 1996, 34, 131–145. [Google Scholar] [CrossRef]
- Benkerroum, N. Antimicrobial peptides generated from milk proteins: A survey and prospects for application in the industry—A review. Int. J. Dairy Technol. 2010, 63, 320–338. [Google Scholar] [CrossRef]
- McPhee, J.B.; Hancock, R.W. Function and therapeutic potential of host defense peptides. J. Peptide Sci. 2005, 11, 677–687. [Google Scholar] [CrossRef]
- Hammami, R.; Zouhir, A.; Ben Hamida, J.; Fliss, I. BACTIBASE: A new web-accessible database for bacteriocin characterization. BMC Microbiol. 2007, 7, 89. [Google Scholar] [CrossRef]
- Steinstraesser, L.; Kraneburg, U.; Jacobson, F.; Al-Benna, S. Host defense peptides and their antimicrobial-immunomodulatory duality. Immunobiology 2011, 216, 322–333. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Harris, F.; Dennison, S.R.; Phoenix, D.A. Anionic antimicrobial peptides from eukaryotic organisms. Curr. Protein Pept. Sci. 2009, 10, 585–606. [Google Scholar] [CrossRef]
- Wakabayashi, H.; Takase, M.; Tomita, M. Lactoferricin derived from milk protein lactoferrin. Curr. Pharm. Des. 2003, 9, 1277–1287. [Google Scholar] [CrossRef]
- Guruprasad, K.; Reddy, B.V.B.; Pandit, M.W. Correlation between stability of a protein and its dipeptide composition—A novel-approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990, 4, 155–161. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- CAMP. Available online: http://www.camp.bicnirrh.res.in/ (accessed from 10 February to 30 April 2014).
- Korhonen, H.; Pihlanto, A. Bioactive peptides: Production and functionality. Int. Dairy J. 2006, 16, 945–960. [Google Scholar] [CrossRef]
- López-Expósito, I.; Minervini, F.; Amigo, L.; Recio, I. Identification of antibacterial peptides from bovine kappa-casein. J. Food Prot. 2006, 69, 2992–2997. [Google Scholar]
- McCann, K.B.; Shiell, B.J.; Michalski, W.P.; Lee, A.; Wan, J.; Roginski, H.; Coventry, M.J. Isolation and characterisation of a novel antibacterial peptide from bovine αS1-casein. Int. Dairy J. 2006, 16, 316–323. [Google Scholar] [CrossRef]
- Pellegrini, A.; Dettling, C.; Thomas, U.; Hunziker, P. Isolation and identification of four bactericidal domains in the bovine β-lactoglobulin. Biochim. Biophys. Acta 2001, 1526, 131–140. [Google Scholar]
- Lignitto, L.; Segato, S.; Balzan, S.; Cavatorta, V.; Oulahal, N.; Sforza, S.; Degraeve, P.; Galaverna, P.; Novelli, E. Preliminary investigation on the presence of peptides inhibiting the growth of Listeria innocua and Listeria monocytogenes in Asiago d’Allevo cheese. Dairy Sci. Technol. 2012, 92, 297–308. [Google Scholar] [CrossRef]
- Meira, S.M.M.; Daroit, D.J.; Helfer, V.E.; Corrêa, A.P.F.; Jéferson, S.; Carro, S.; Brandelli, A. Bioactive peptides in water-soluble extracts of ovine cheeses from Southern Brazil and Uruguay. Food Res. Int. 2012, 48, 322–329. [Google Scholar] [CrossRef]
- Rizzello, C.G.; Gobbetti, L.M.; Carbonara, T.; de Bari, M.D.; Zambonin, P.G. Antibacterial activities of peptides from the water-soluble extracts of Italian cheese varieties. J. Dairy Sci. 2005, 88, 2348–2360. [Google Scholar] [CrossRef]
- Agyei, D.; Danquah, M.K. Rethinking food-derived bioactive peptides for antimicrobial and immunomodulatory activities. Trends Food Sci. Technol. 2012, 23, 62–69. [Google Scholar] [CrossRef]
- Agyei, D.; Danquah, M.K. Industrial-scale manufacturing of pharmaceutical-grade bioactive peptides. Biotechnol. Adv. 2011, 29, 272–277. [Google Scholar] [CrossRef]
- PMAP. Available online: http://www.proteolysis.org/proteases (accessed on 7 January 2014).
- PeptideCutter. Available online: http://www.expasy.org/tools/peptidecutter/ (accessed on 7 January 2014).
- Wang, G.S.; Li, X.; Wang, Z. APD2: The updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009, 37, D933–D937. [Google Scholar] [CrossRef]
- Hammami, R.; Ben Hamida, J.; Vergoten, G.; Fliss, I. PhytAMP: A database dedicated to antimicrobial plant peptides. Nucleic Acids Res. 2009, 37, D963–D968. [Google Scholar] [CrossRef]
- Lata, S.; Sharma, BK.; Raghava, G.P.S. Analysis and prediction of antibacterial peptides. BMC Bioinform. 2007, 8, 263. [Google Scholar] [CrossRef]
- Torrent, M.; di Tommaso, P.; Pulido, D.; Nogués, M.V.; Notredame, C.; Boix, E.; Andreu, D. AMPA: An automated web server for prediction of protein antimicrobial regions. Bioinformatics 2012, 28, 130–131. [Google Scholar] [CrossRef]
- Waghu, F.H.; Gopi, L.; Barai, R.S.; Ramteke, P.; Nizami, B.; Idicula-Thomas, S. CAMP: Collection of sequences and structures of antimicrobial peptides. Nucleic Acids Res. 2014, 42, D1154–D1158. [Google Scholar] [CrossRef]
- Peptide Property Calculator. Available online: http://www.innovagen.se/custom-peptide-synthesis/peptide-property-calculator/peptide-property-calculator.asp (accessed from 7 January to 28 April 2014).
- Marshall, S.H.; Arenas, G. Antimicrobial peptides: A natural alternative to chemical antibiotics and a potential for applied biotechnology. Elektron. J. Biotechnol. 2003, 6, 271–282. [Google Scholar]
- Strøm, M.B.; Haug, B.E.; Skar, M.L.; Stensen, W.; Stiberg, T.; Svendsen, J.S. The pharmacophore of short cationic antibacterial peptides. J. Med. Chem. 2003, 46, 1567–1570. [Google Scholar] [CrossRef]
- López-Expósito, I.; Gómez-Ruiz, J.A.; Amigo, L.; Recio, I. Identification of antibacterial peptides from ovine αS2-casein. Int. Dairy J. 2006, 16, 1072–1080. [Google Scholar] [CrossRef]
- Meng, S.; Xu, H.; Wang, F. Research advances of antimicrobial peptides and applications in food industry and agriculture. Curr. Protein Pept. Sci. 2010, 11, 264–273. [Google Scholar] [CrossRef]
- Vignes, R. Dimethyl Sulfoxide (DMSO)—A “new” clean, unique, superior solvent. In American Chemical Society Annual Meeting, Washington, DC, USA; 2000; pp. 1–20. [Google Scholar]
- ProtParam. Available online: http://web.expasy.org/protparam/ (accessed from 7 January to 28 February 2014).
- APD2: Antimicrobial Peptide Calculator and Predictor. Available online: http://aps.unmc.edu/AP/prediction/prediction_main.php (accessed on 30 April 2014).
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Dziuba, B.; Dziuba, M. New Milk Protein-Derived Peptides with Potential Antimicrobial Activity: An Approach Based on Bioinformatic Studies. Int. J. Mol. Sci. 2014, 15, 14531-14545. https://doi.org/10.3390/ijms150814531
Dziuba B, Dziuba M. New Milk Protein-Derived Peptides with Potential Antimicrobial Activity: An Approach Based on Bioinformatic Studies. International Journal of Molecular Sciences. 2014; 15(8):14531-14545. https://doi.org/10.3390/ijms150814531
Chicago/Turabian StyleDziuba, Bartłomiej, and Marta Dziuba. 2014. "New Milk Protein-Derived Peptides with Potential Antimicrobial Activity: An Approach Based on Bioinformatic Studies" International Journal of Molecular Sciences 15, no. 8: 14531-14545. https://doi.org/10.3390/ijms150814531




