Elucidating Polypharmacological Mechanisms of Polyphenols by Gene Module Profile Analysis
Abstract
:1. Introduction
2. Results and Discussion

| Drugs | Therapeutic Uses | Targets | References |
|---|---|---|---|
| Imatinib | Antineoplastic Agents | Platelet-derived growth factor receptor a | [33] |
| Proto-oncogene tyrosine-protein kinase ABL1 a | [34] | ||
| Mast/stem cell growth factor receptor a | [35] | ||
| Raloxifene | Antihypocalcemic Agents | Estrogen receptor a | [36] |
| Iloprost | Antihypertensive Agents | Prostaglandin E2 receptor, EP2 subtype b | [37] |
| cAMP-specific 3',5'-cyclic phosphodiesterase a | [38] | ||
| Prostacyclin receptor c | [37] | ||
| Cisapride | Anti-Ulcer Agents | 5-Hydroxytryptamine 4 receptor | - |
| Gastrointestinal Agents | |||
| Prokinetic Agents | |||
| Fluticasone | Anti-inflammatory Agents | Glucocorticoid receptor a | [39] |
| Diethylstilbestrol | Antineoplastic Agents | Estrogen receptor a | [36] |
| Finasteride | Anti-baldness Agents | Steroid-5-alpha reductase a | [40] |
| Antihyperplasia Agents | |||
| Sulindac sulfide | Rheumatoid arthritis | - | - |
| Prednisone | Anti-inflammatory Agents | Glucocorticoid receptor a | [39] |
| Antineoplastic Agents | |||
| Estradiol | Anti-menopausal Agents | Estrogen receptor a | [36] |
| Anticholesteremic Agents | |||
| Dydrogesterone | Progesterones | Progesterone receptor |
| Drugs | Therapeutic Uses | Targets | References |
|---|---|---|---|
| Tolazoline | Adrenergic alpha-Antagonists | Alpha adrenergic receptor | - |
| Antihypertensive Agents | |||
| Vasodilator Agents | |||
| Tamoxifen | Antineoplastic Agents | Estrogen receptor a | [41] |
| Bone Density Conservation Agents | |||
| Finasteride | Anti-baldness Agents | Steroid-5-alpha reductase | - |
| Antihyperplasia Agents | |||
| Skin and Mucous Membrane Agents | |||
| Sulindac sulfide | Rheumatoid arthritis | - | - |
| Iloprost | Antihypertensive Agents | Prostaglandin E2 receptor, EP2 subtype | - |
| cAMP-specific 3',5'-cyclic phosphodiesterase a | [42] | ||
| Prostacyclin receptor | - | ||
| Raloxifene | Antihypocalcemic Agents | Estrogen receptor a | [41] |
| Bone Density Conservation Agents | |||
| Apomorphine | Antiparkinson Agents | Dopamine receptor a | [43] |
| Adrenergic receptors | - | ||
| 5-Hydroxytryptamine receptor a | [43] | ||
| Fluticasone | Anti-inflammatory Agents | Glucocorticoid receptor | - |
| Tocainide | Anti-Arrhythmia Agents | Sodium channel protein type 5 subunit alpha a | [44] |
| Drugs | Therapeutic Uses | Targets | References |
|---|---|---|---|
| Reserpine | Antihypertensive Agents | Synaptic vesicular amine transporter | - |
| Antipsychotic Agents | |||
| Mercaptopurine | Antineoplastic Agents | Hypoxanthine-guanine phosphoribosyltransferase | - |
| Immunosuppressive Agents | |||
| Niclosamide | Antiparasitic Agents | - | - |
| Daunorubicin | Antineoplastic Agents | DNA topoisomerase | - |
| Terfenadine | Anti-Allergic Agents | Histamine H1 receptor | - |
| Antiarrhythmic Agents | Potassium voltage-gated channel subfamily H member 2 a | [45] | |
| Muscarinic acetylcholine receptor M3 | - | ||
| Fluphenazine | Antipsychotic Agents | Dopamine receptor | - |
| Dipyridamole | Vasodilator Agents | Adenosine deaminase | - |
| cGMP-specific 3',5'-cyclic phosphodiesterase a | [46] | ||
| Rescinnamine | Antihypertensive Agents | Angiotensin-converting enzyme a | [47] |
| Trifluoperazine | Antipsychotic Agents | Dopamine receptor | - |
| Metixene | Antiparkinson Agents | Muscarinic acetylcholine receptor | - |
| Drugs | Therapeutic Uses | Targets | References |
|---|---|---|---|
| Letrozole | Antineoplastic Agents | Cytochrome P450 19A1 a | [48] |
| Triprolidine | Anti-Allergic Agents | Histamine H1 receptor | |
| Pindolol | Antihypertensive Agents | Adrenergic receptor | - |
| Vasodilator Agents | 5-hydroxytryptamine receptor | - | |
| Norfloxacin | Anti-Bacterial Agents | DNA topoisomerase 2-alpha a | [48] |
| Prilocaine | Anesthetics | Sodium channel protein type 5 subunit alpha | - |
| Estradiol | Anti-menopausal Agents | Estrogen receptor a | [49] |
| Anticholesteremic Agents | |||
| Doxycycline | Anti-Bacterial Agents | 30S ribosomal protein | - |
| Bendroflumethiazide | Antihypertensive Agents | Solute carrier family 12 member 3 | - |
| Calcium-activated potassium channel subunit alpha 1 | - | ||
| Carbonic anhydrase | - | ||
| Theophylline | Bronchodilator Agents | Adenosine A1 receptor | - |
| Vasodilator Agents | cGMP-specific 3',5'-cyclic phosphodiesterase a | [29] | |
| Naltrexone | Anti-craving Agents | Opioid receptor a | [50] |
3. Experimental
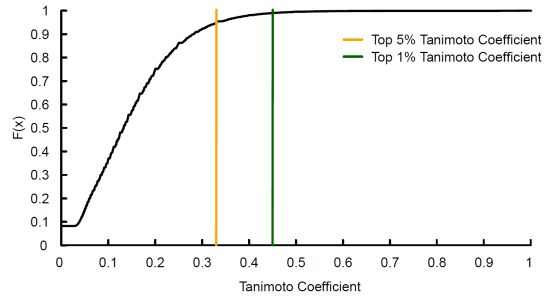
3.1. Tanimoto Coefficient Calculation
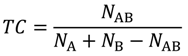
3.2. Drug Target Enrichment
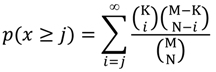
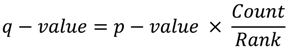
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Halliwell, B.; Gutteridge, J. Free Radicals in Biology and Medicine; Oxford University Press: New York, NY, USA, 2007. [Google Scholar]
- Rice-Evans, C.A.; Miller, N.J.; Paganga, G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radic. Biol. Med. 1996, 20, 933–956. [Google Scholar] [CrossRef]
- Zhang, H.Y. Structure-activity relationships and rational design strategies for radical-scavenging antioxidants. Curr. Comput. Aided Drug Des. 2005, 1, 257–273. [Google Scholar] [CrossRef]
- Fernández-Arroyo, S.; Herranz-López, M.; Beltrán-Debón, R.; Borrás-Linares, I.; Barrajón-Catalán, E.; Joven, J.; Fernández-Gutiérrez, A.; Segura-Carretero, A.; Micol, V. Bioavailability study of a polyphenol-enriched extract from Hibiscus Sabdariffa in rats and associated antioxidant status. Mol. Nutr. Food Res. 2012, 56, 1590–1595. [Google Scholar] [CrossRef]
- Melton, L. The antioxidant myth: a medical fairy tale. New Sci. 2006, 2563, 40–43. [Google Scholar]
- Pun, P.B.; Gruber, J.; Tang, S.Y.; Schaffer, S.; Ong, R.L.; Fong, S.; Ng, L.F.; Cheah, I.; Halliwell, B. Ageing in nematodes: Do antioxidants extend lifespan in Caenorhabditis elegans? Biogerontology 2010, 11, 17–30. [Google Scholar] [CrossRef]
- Stanner, S.A.; Hughes, J.; Kelly, C.N.; Buttriss, J. A review of the epidemiological evidence for the “antioxidant hypothesis”. Public Health Nutr. 2004, 7, 407–422. [Google Scholar]
- Hollman, P.C.; Cassidy, A.; Comte, B.; Heinonen, M.; Richelle, M.; Richling, E.; Serafini, M.; Scalbert, A.; Sies, H.; Vidry, S. The biological relevance of direct antioxidant effects of polyphenols for cardiovascular health in humans is not established. J. Nutr. 2011, 141, 989S–1009S. [Google Scholar] [CrossRef]
- Halliwell, B. Free radicals and antioxidants—Quo vadis? Trends Pharmacol. Sci. 2011, 32, 125–130. [Google Scholar] [CrossRef]
- Halliwell, B.; Rafter, J.; Jenner, A. Health promotion by flavonoids, tocopherols, tocotrienols, and other phenols: direct or indirect effects? Antioxidant or not? Am. J. Clin. Nutr. 2005, 81, 268S–276S. [Google Scholar]
- Zhang, H.Y.; Chen, L.L.; Li, X.J.; Zhang, J. Evolutionary inspirations for drug discovery. Trends Pharmacol. Sci. 2010, 31, 443–448. [Google Scholar] [CrossRef]
- Ji, H.F.; Li, X.J.; Zhang, H.Y. Natural products and drug discovery. Can thousands of years of ancient medical knowledge lead us to new and powerful drug combinations in the fight against cancer and dementia? EMBO Rep. 2009, 10, 194–200. [Google Scholar] [CrossRef]
- Middleton, E.; Kandaswami, C.; Theoharides, T.C. The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease, and cancer. Pharmacol. Rev. 2000, 52, 673–751. [Google Scholar]
- Yue, R.; Shan, L.; Yang, X.; Zhang, W. Approaches to target profiling of natural products. Curr. Med. Chem. 2012, 19, 3841–3855. [Google Scholar] [CrossRef]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef]
- Iorio, F.; Bosotti, R.; Scacheri, E.; Belcastro, V.; Mithbaokar, P.; Ferriero, R.; Murino, L.; Tagliaferri, R.; Brunetti-Pierri, N.; Isacchi, A.; et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc. Natl. Acad. Sci. USA 2010, 107, 14621–14626. [Google Scholar] [CrossRef]
- Qu, X.A.; Rajpal, D.K. Applications of Connectivity Map in drug discovery and development. Drug Discov. Today 2012, 17, 1289–1298. [Google Scholar] [CrossRef]
- Hochreiter, S.; Bodenhofer, U.; Heusel, M.; Mayr, A.; Mitterecker, A.; Kasim, A.; Khamiakova, T.; van Sanden, S.; Lin, D.; Talloen, W.; et al. A. FABIA: factor analysis for bicluster acquisition. Bioinformatics 2010, 26, 1520–1527. [Google Scholar] [CrossRef]
- Xiong, M.; Li, B.; Zhu, Q.; Wang, Y.X.; Zhang, H.Y. Identification of transcription factors for drug-associated gene modules and biomedical implications. Bioinformatics 2013, 30, 305–309. [Google Scholar]
- Wishart, D.S. DrugBank and its relevance to pharmacogenomics. Pharmacogenomics 2008, 9, 1155–1162. [Google Scholar] [CrossRef]
- Zhu, F.; Shi, Z.; Qin, C.; Tao, L.; Liu, X.; Xu, F.; Zhang, L.; Song, Y.; Zhang, J.; Han, B.; et al. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012, 40, D1128–D1136. [Google Scholar] [CrossRef]
- Stelzer, G.; Inger, A.; Olender, T.; Iny-Stein, T.; Dalah, I.; Harel, A.; Safran, M.; Lancet, D. GeneDecks: Paralog hunting and gene-set distillation with GeneCards annotation. OMICS 2009, 13, 477–487. [Google Scholar] [CrossRef]
- Chen, Z.Y.; Peng, C.; Jiao, R.; Wong, Y.M.; Yang, N.; Huang, Y. Anti-hypertensive nutraceuticals and functional foods. J. Agric. Food Chem. 2009, 57, 4485–4499. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, W.; Sun, L.; Yu, H.; Ni, Q.X.; Risch, H.A.; Gao, Y.T. Green tea drinking and risk of pancreatic cancer: A large-scale, population-based case-control study in urban Shanghai. Cancer Epidemiol. 2012, 36, e354–e358. [Google Scholar] [CrossRef]
- Kurahashi, N.; Sasazuki, S.; Iwasaki, M.; Inoue, M.; Tsugane, S.; Grp, J.S. Green tea consumption and prostate cancer risk in Japanese men: A prospective study. Am. J. Epidemiol. 2008, 167, 71–77. [Google Scholar]
- Trichopoulou, A.; Lagiou, P.; Kuper, H.; Trichopoulos, D. Cancer and Mediterranean dietary traditions. Cancer Epidemiol. Biomark. Prev. 2000, 9, 869–873. [Google Scholar]
- Erlund, I. Review of the flavonoids quercetin, hesperetin, and naringenin. Dietary sources, bioactivities, and epidemiology. Nutr. Res. 2004, 24, 851–874. [Google Scholar] [CrossRef]
- Bhat, K.P.; Pezzuto, J.M. Cancer chemopreventive activity of resveratrol. Ann. N. Y. Acad. Sci. 2002, 957, 210–229. [Google Scholar] [CrossRef]
- Beretz, A.; Anton, R.; Stoclet, J.C. Flavonoid compounds are potent inhibitors of cyclic AMP phosphodiesterase. Experientia 1978, 34, 1054–1055. [Google Scholar] [CrossRef]
- Kuiper, G.G.; Lemmen, J.G.; Carlsson, B.; Corton, J.C.; Safe, S.H.; van der Saag, P.T.; van der Burg, B.; Gustafsson, J.A. Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor beta. Endocrinology 1998, 139, 4252–4263. [Google Scholar]
- Goodin, M.G.; Fertuck, K.C.; Zacharewski, T.R.; Rosengren, R.J. Estrogen receptor-mediated actions of polyphenolic catechins in vivo and in vitro. Toxicol. Sci. 2002, 69, 354–361. [Google Scholar] [CrossRef]
- Baur, J.A.; Mai, A. Revelations into resveratrol’s mechanism. Nat. Med. 2012, 18, 500–501. [Google Scholar] [CrossRef]
- Yan, G.R.; Xiao, C.L.; He, G.W.; Yin, X.F.; Chen, N.P.; Cao, Y.; He, Q.Y. Global phosphoproteomic effects of natural tyrosine kinase inhibitor, genistein, on signaling pathways. Proteomics 2010, 10, 976–986. [Google Scholar]
- Lin, M.Q.; den Dulk-Ras, A.; Hooykaas, P.J.J.; Rikihisa, Y. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol. 2007, 9, 2644–2657. [Google Scholar] [CrossRef]
- Packer, A.I.; Hsu, Y.C.; Besmer, P.; Bachvarova, R.F. The ligand of the c-kit receptor promotes oocyte growth. Dev. Biol. 1994, 161, 194–205. [Google Scholar] [CrossRef]
- Rickard, D.J.; Monroe, D.G.; Ruesink, T.J.; Khosla, S.; Riggs, B.L.; Spelsberg, T.C. Phytoestrogen genistein acts as an estrogen agonist on human osteoblastic cells through estrogen receptors alpha and beta. J. Cell Biochem. 2003, 89, 633–646. [Google Scholar] [CrossRef]
- Hermenegildo, C.; Oviedo, P.J.; Garcia-Perez, M.A.; Tarin, J.J.; Cano, A. Effects of phytoestrogens genistein and daidzein on prostacyclin production by human endothelial cells. J. Pharmacol. Exp. Ther. 2005, 315, 722–728. [Google Scholar] [CrossRef]
- Shih, C.-H.; Lin, L.-H.; Lai, Y.-H.; Lai, C.-Y.; Han, C.-Y.; Chen, C.-M.; Ko, W.-C. Genistein, a competitive PDE1-4 inhibitor, may bind on high-affinity rolipram binding sites of brain cell membranes and then induce gastrointestinal adverse effects. Eur. J. Pharmacol. 2010, 643, 113–120. [Google Scholar] [CrossRef]
- Nishizaki, Y.; Ishimoto, Y.; Hotta, Y.; Hosoda, A.; Yoshikawa, H.; Akamatsu, M.; Tamura, H. Effect of flavonoids on androgen and glucocorticoid receptors based on in vitro reporter gene assay. Bioorg. Med. Chem. Lett. 2009, 19, 4706–4710. [Google Scholar] [CrossRef]
- Ye, L.; Su, Z.J.; Ge, R.S. Inhibitors of testosterone biosynthetic and metabolic activation enzymes. Molecules 2011, 16, 9983–10001. [Google Scholar] [CrossRef]
- Maggiolini, M.; Bonofiglio, D.; Marsico, S.; Panno, M.L.; Cenni, B.; Picard, D.; Ando, S. Estrogen receptor alpha mediates the proliferative but not the cytotoxic dose-dependent effects of two major phytoestrogens on human breast cancer cells. Mol. Pharmacol. 2001, 60, 595–602. [Google Scholar]
- Lines, T.C.; Ono, M. FRS 1000, an extract of red onion peel, strongly inhibits phosphodiesterase 5A (PDE 5A). Phytomedicine 2006, 13, 236–239. [Google Scholar] [CrossRef]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B.; et al. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef]
- Wallace, C.H.R.; Baczko, I.; Jones, L.; Fercho, M.; Light, P.E. Inhibition of cardiac voltage-gated sodium channels by grape polyphenols. Br. J. Pharmacol. 2006, 149, 657–665. [Google Scholar] [CrossRef]
- Granados-Soto, V.; Arguelles, C.F.; Ortiz, M.I. The peripheral antinociceptive with activation of effect of resveratrol is associated potassium channels. Neuropharmacology 2002, 43, 917–923. [Google Scholar] [CrossRef]
- Park, S.J.; Ahmad, F.; Philp, A.; Baar, K.; Williams, T.; Luo, H.; Ke, H.; Rehmann, H.; Taussig, R.; Brown, A.L.; et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell 2012, 148, 421–433. [Google Scholar] [CrossRef]
- Melzig, M.F.; Escher, F. Induction of neutral endopeptidase and anglotensin-converting enzyme activity of SK-N-SH cells in vitro by quercetin and resveratrol. Pharmazie 2002, 57, 556–558. [Google Scholar]
- Davis, A.P.; King, B.L.; Mockus, S.; Murphy, C.G.; Saraceni-Richards, C.; Rosenstein, M.; Wiegers, T.; Mattingly, C.J. The comparative toxicogenomics database: Update 2011. Nucleic Acids Res. 2011, 39, D1067–D1072. [Google Scholar] [CrossRef]
- Damianaki, A.; Bakogeorgou, E.; Kampa, M.; Notas, G.; Hatzoglou, A.; Panagiotou, S.; Gemetzi, C.; Kouroumalis, E.; Martin, P.-M.; Castanas, E. Potent inhibitory action of red wine polyphenols on human breast cancer cells. J. Cell Biochem. 2000, 78, 429–441. [Google Scholar] [CrossRef]
- Katavic, P.L.; Lamb, K.; Navarro, H.; Prisinzano, T.E. Flavonoids as opioid receptor ligands: Identification and preliminary structure-activity relationships. J. Nat. Prod. 2007, 70, 1278–1282. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995, 57, 289–300. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, B.; Xiong, M.; Zhang, H.-Y. Elucidating Polypharmacological Mechanisms of Polyphenols by Gene Module Profile Analysis. Int. J. Mol. Sci. 2014, 15, 11245-11254. https://doi.org/10.3390/ijms150711245
Li B, Xiong M, Zhang H-Y. Elucidating Polypharmacological Mechanisms of Polyphenols by Gene Module Profile Analysis. International Journal of Molecular Sciences. 2014; 15(7):11245-11254. https://doi.org/10.3390/ijms150711245
Chicago/Turabian StyleLi, Bin, Min Xiong, and Hong-Yu Zhang. 2014. "Elucidating Polypharmacological Mechanisms of Polyphenols by Gene Module Profile Analysis" International Journal of Molecular Sciences 15, no. 7: 11245-11254. https://doi.org/10.3390/ijms150711245
APA StyleLi, B., Xiong, M., & Zhang, H.-Y. (2014). Elucidating Polypharmacological Mechanisms of Polyphenols by Gene Module Profile Analysis. International Journal of Molecular Sciences, 15(7), 11245-11254. https://doi.org/10.3390/ijms150711245