miR-137: A New Player in Schizophrenia
Abstract
:1. Introduction
2. Expression and Epigenetic Regulation of miR-137
3. The Function of miR-137 in the Nervous System
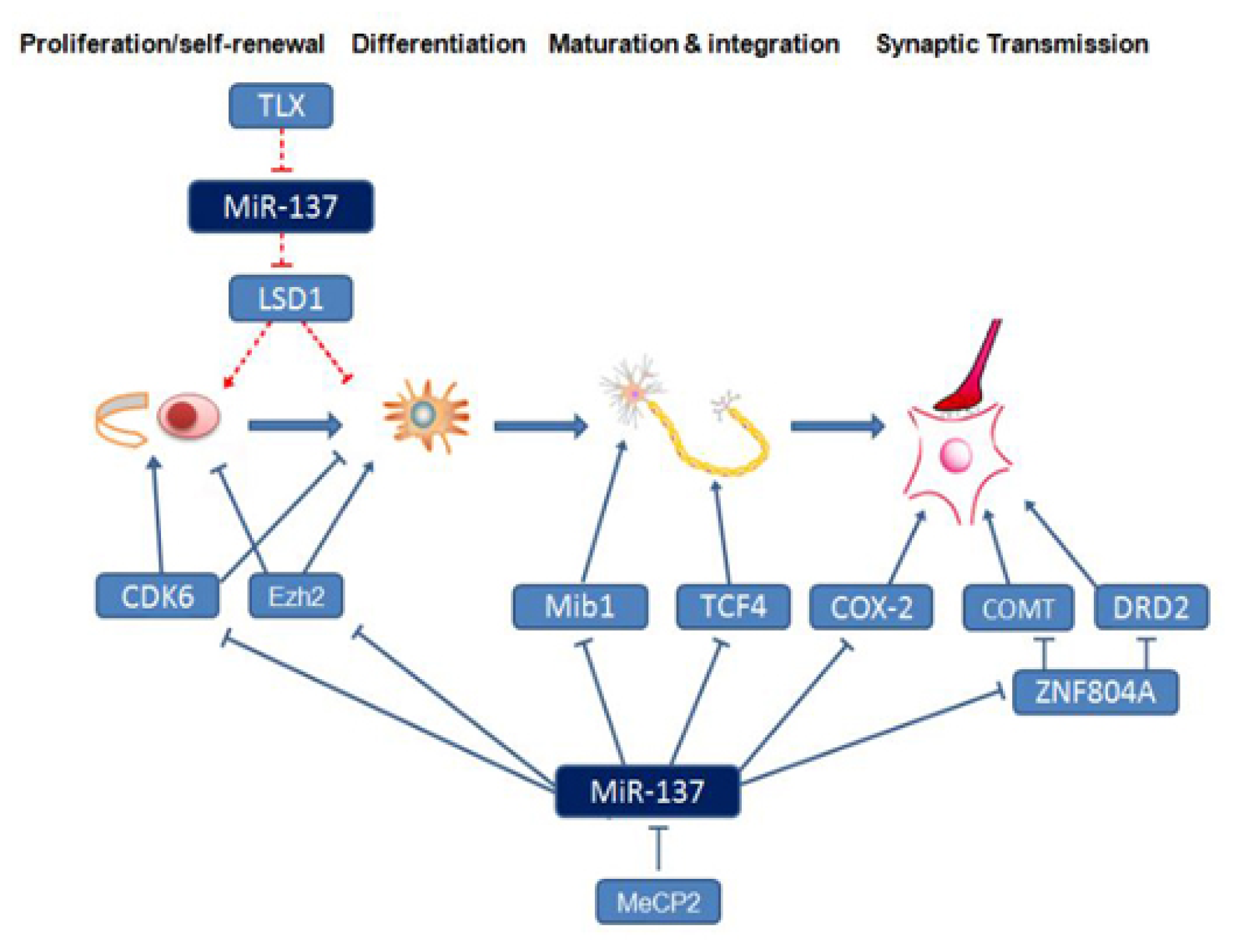
4. Neural Development- and Schizophrenia-Related Downstream Target Genes and Signaling Pathways Regulated by miR-137
5. Correlations between miR-137 Single Nucleotide Polymorphisms and Schizophrenia
6. Summary and Prospects
Acknowledgments
Conflicts of Interest
References
- Allen, N.C.; Bagade, S.; McQueen, M.B.; Ioannidis, J.P.; Kavvoura, F.K.; Khoury, M.J.; Tanzi, R.E.; Bertram, L. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: The SzGene database. Nat. Genet 2008, 40, 827–834. [Google Scholar]
- Cannon, T.D. Abnormalities of brain structure and function in schizophrenia: implications for aetiology and pathophysiology. Ann. Med 1996, 28, 533–539. [Google Scholar]
- Murchison, E.P.; Partridge, J.F.; Tam, O.H.; Cheloufi, S.; Hannon, G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA 2005, 102, 12135–12140. [Google Scholar]
- Wang, Y.; Medvid, R.; Melton, C.; Jaenisch, R.; Blelloch, R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet 2007, 39, 380–385. [Google Scholar]
- Stark, K.L.; Xu, B.; Bagchi, A.; Lai, W.S.; Liu, H.; Hsu, R.; Wan, X.; Pavlidis, P.; Mills, A.A.; Karayiorgou, M.; et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet 2008, 40, 751–760. [Google Scholar]
- Sewell, R.A.; Perry, E.B., Jr.; Karper, L.P.; Bell, M.D.; Lysaker, P.; Goulet, J.L.; Brenner, L.; Erdos, J.; d’Souza, D.C.; Seibyl, J.P.; et al. Clinical significance of neurological soft signs in schizophrenia: Factor analysis of the Neurological Evaluation Scale. Schizophr. Res 2010, 124, 1–12. [Google Scholar]
- Perkins, D.O.; Jeffries, C.D.; Jarskog, L.F.; Thomson, J.M.; Woods, K.; Newman, M.A.; Parker, J.S.; Jin, J.; Hammond, S.M. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol 2007, 8, R27. [Google Scholar]
- Xu, B.; Karayiorgou, M.; Gogos, J.A. MicroRNAs in psychiatric and neurodevelopmental disorders. Brain Res 2010, 1338, 78–88. [Google Scholar]
- Guella, I.; Sequeira, A.; Rollins, B.; Morgan, L.; Torri, F.; van Erp, T.G.; Myers, R.M.; Barchas, J.D.; Schatzberg, A.F.; Watson, S.J.; et al. Analysis of miR-137 expression and rs1625579 in dorsolateral prefrontal cortex. J. Psychiatr. Res 2013, 47, 1215–1221. [Google Scholar]
- Smrt, R.D.; Szulwach, K.E.; Pfeiffer, R.L.; Li, X.; Guo, W.; Pathania, M.; Teng, Z.Q.; Luo, Y.; Peng, J.; Bordey, A.; et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 2010, 28, 1060–1070. [Google Scholar]
- Kunej, T.; Godnic, I.; Horvat, S.; Zorc, M.; Calin, G.A. Cross talk between microRNA and coding cancer genes. Cancer J 2012, 18, 223–231. [Google Scholar]
- Sun, G.; Ye, P.; Murai, K.; Lang, M.F.; Li, S.; Zhang, H.; Li, W.; Fu, C.; Yin, J.; Wang, A.; et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat. Commun 2011, 2, 529. [Google Scholar]
- Silber, J.; Lim, D.A.; Petritsch, C.; Persson, A.I.; Maunakea, A.K.; Yu, M.; Vandenberg, S.R.; Ginzinger, D.G.; James, C.D.; Costello, J.F.; et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med 2008, 6, 14. [Google Scholar]
- Lewis, J.D.; Meehan, R.R.; Henzel, W.J.; Maurer-Fogy, I.; Jeppesen, P.; Klein, F.; Bird, A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 1992, 69, 905–914. [Google Scholar]
- Szulwach, K.E.; Li, X.; Smrt, R.D.; Li, Y.; Luo, Y.; Lin, L.; Santistevan, N.J.; Li, W.; Zhao, X.; Jin, P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol 2010, 189, 127–141. [Google Scholar]
- Jiang, K.; Ren, C.; Nair, V.D. MicroRNA-137 represses Klf4 and Tbx3 during differentiation of mouse embryonic stem cells. Stem Cell Res 2013, 11, 1299–1313. [Google Scholar]
- Tarantino, C.; Paolella, G.; Cozzuto, L.; Minopoli, G.; Pastore, L.; Parisi, S.; Russo, T. miRNA 34a, 100, and 137 modulate differentiation of mouse embryonic stem cells. FASEB J 2010, 24, 3255–3263. [Google Scholar]
- Breder, C.D.; Dewitt, D.; Kraig, R.P. Characterization of inducible cyclooxygenase in rat brain. J. Comp. Neurol 1995, 355, 296–315. [Google Scholar]
- Akaneya, Y. The remarkable mechanism of prostaglandin E2 on synaptic plasticity. Gene Regul. Syst. Biol 2007, 1, 83–89. [Google Scholar]
- Sang, N.; Zhang, J.; Marcheselli, V.; Bazan, N.G.; Chen, C. Postsynaptically synthesized prostaglandin E2 (PGE2) modulates hippocampal synaptic transmission via a presynaptic PGE2 EP2 receptor. J. Neurosci 2005, 25, 9858–9870. [Google Scholar]
- Guilmatre, A.; Dubourg, C.; Mosca, A.L.; Legallic, S.; Goldenberg, A.; Drouin-Garraud, V.; Layet, V.; Rosier, A.; Briault, S.; Bonnet-Brilhault, F.; et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch. Gen. Psychiatry 2009, 66, 947–956. [Google Scholar]
- Ripke, S.; Sanders, A.; Kendler, K.; Levinson, D.; Sklar, P.; Holmans, P. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet 2011, 43, 969–976. [Google Scholar]
- Kwon, E.; Wang, W.; Tsai, L.H. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Mol. Psychiatry 2011, 18, 11–12. [Google Scholar]
- Brzozka, M.M.; Radyushkin, K.; Wichert, S.P.; Ehrenreich, H.; Rossner, M.J. Cognitive and sensorimotor gating impairments in transgenic mice overexpressing the schizophrenia susceptibility gene Tcf4 in the brain. Biol. Psychiatry 2010, 68, 33–40. [Google Scholar]
- Green, E.K.; Grozeva, D.; Jones, I.; Jones, L.; Kirov, G.; Caesar, S.; Gordon-Smith, K.; Fraser, C.; Forty, L.; Russell, E.; et al. The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol. Psychiatry 2010, 15, 1016–1022. [Google Scholar]
- Dao, D.T.; Mahon, P.B.; Cai, X.; Kovacsics, C.E.; Blackwell, R.A.; Arad, M.; Shi, J.; Zandi, P.P.; O’Donnell, P.; Knowles, J.A.; et al. Mood disorder susceptibility gene CACNA1C modifies mood-related behaviors in mice and interacts with sex to influence behavior in mice and diagnosis in humans. Biol. Psychiatry 2010, 68, 801–810. [Google Scholar]
- Bhat, S.; Dao, D.T.; Terrillion, C.E.; Arad, M.; Smith, R.J.; Soldatov, N.M.; Gould, T.D. CACNA1C (Cav1.2) in the pathophysiology of psychiatric disease. Prog. Neurobiol 2012, 99, 1–14. [Google Scholar]
- Kim, A.H.; Parker, E.K.; Williamson, V.; McMichael, G.O.; Fanous, A.H.; Vladimirov, V.I. Experimental validation of candidate schizophrenia gene ZNF804A as target for hsa-miR-137. Schizophr. Res 2012, 141, 60–64. [Google Scholar]
- Girgenti, M.J.; LoTurco, J.J.; Maher, B.J. ZNF804a regulates expression of the schizophrenia-associated genes PRSS16, COMT, PDE4B, and DRD2. PLoS One 2012, 7, e32404. [Google Scholar]
- Liu, L.; Yuan, G.; Cheng, Z.; Zhang, G.; Liu, X.; Zhang, H. Identification of the mRNA expression status of the dopamine d2 receptor and dopamine transporter in peripheral blood lymphocytes of schizophrenia patients. PLoS One 2013, 8, e75259. [Google Scholar]
- Wright, C.; Turner, J.A.; Calhoun, V.D.; Perrone-Bizzozero, N. Potential impact of miR-137 and its targets in schizophrenia. Front. Genet 2013, 4, 58. [Google Scholar]
- Skaper, S.D. Neuronal growth-promoting and inhibitory cues in neuroprotection and neuroregeneration. Methods Mol. Biol 2012, 846, 13–22. [Google Scholar]
- Yang, Y.; Calakos, N. Presynaptic long-term plasticity. Front. Synaptic Neurosci 2013, 5, 8. [Google Scholar]
- Du, J.; Fu, C.; Sretavan, D.W. Eph/ephrin signaling as a potential therapeutic target after central nervous system injury. Curr. Pharm. Des 2007, 13, 2507–2518. [Google Scholar]
- Nikolov, D.B.; Xu, K.; Himanen, J.P. Eph/ephrin recognition and the role of Eph/ephrin clusters in signaling initiation. Biochim. Biophys. Acta 2013, 1834, 2160–2165. [Google Scholar]
- Guan, F.; Zhang, B.; Yan, T.; Li, L.; Liu, F.; Li, T.; Feng, Z.; Liu, X.; Li, S. MIR137 gene and target gene CACNA1C of miR-137 contribute to schizophrenia susceptibility in Han Chinese. Schizophr. Res 2014, 152, 97–104. [Google Scholar]
- Arinami, T.; Ohtsuki, T.; Ishiguro, H.; Ujike, H.; Tanaka, Y.; Morita, Y.; Mineta, M.; Takeichi, M.; Yamada, S.; Imamura, A.; et al. Genomewide high-density SNP linkage analysis of 236 Japanese families supports the existence of schizophrenia susceptibility loci on chromosomes 1p, 14q, and 20p. Am. J. Hum. Genet 2005, 77, 937–944. [Google Scholar]
- Cummings, E.; Donohoe, G.; Hargreaves, A.; Moore, S.; Fahey, C.; Dinan, T.G.; McDonald, C.; O’Callaghan, E.; O’Neill, F.A.; Waddington, J.L.; et al. Mood congruent psychotic symptoms and specific cognitive deficits in carriers of the novel schizophrenia risk variant at MIR-137. Neurosci. Lett 2013, 532, 33–38. [Google Scholar] [Green Version]
- Whalley, H.C.; Papmeyer, M.; Romaniuk, L.; Sprooten, E.; Johnstone, E.C.; Hall, J.; Lawrie, S.M.; Evans, K.L.; Blumberg, H.P.; Sussmann, J.E.; et al. Impact of a microRNA MIR137 Susceptibility Variant on Brain Function in People at High Genetic Risk of Schizophrenia or Bipolar Disorder. Neuropsychopharmacology 2012, 37, 2720–2729. [Google Scholar]
- Green, M.J.; Cairns, M.J.; Wu, J.; Dragovic, M.; Jablensky, A.; Tooney, P.A.; Scott, R.J.; Carr, V.J. Genome-wide supported variant MIR137 and severe negative symptoms predict membership of an impaired cognitive subtype of schizophrenia. Mol. Psychiatry 2013, 18, 774–780. [Google Scholar]
- Lett, T.A.; Chakravarty, M.M.; Felsky, D.; Brandl, E.J.; Tiwari, A.K.; Goncalves, V.F.; Rajji, T.K.; Daskalakis, Z.J.; Meltzer, H.Y.; Lieberman, J.A.; et al. The genome-wide supported microRNA-137 variant predicts phenotypic heterogeneity within schizophrenia. Mol. Psychiatry 2013, 18, 443–450. [Google Scholar]
- Van Erp, T.G.; Guella, I.; Vawter, M.P.; Turner, J.; Brown, G.G.; McCarthy, G.; Greve, D.N.; Glover, G.H.; Calhoun, V.D.; Lim, K.O.; et al. Schizophrenia miR-137 locus risk genotype is associated with dorsolateral prefrontal cortex hyperactivation. Biol. Psychiatry 2014, 75, 398–405. [Google Scholar]
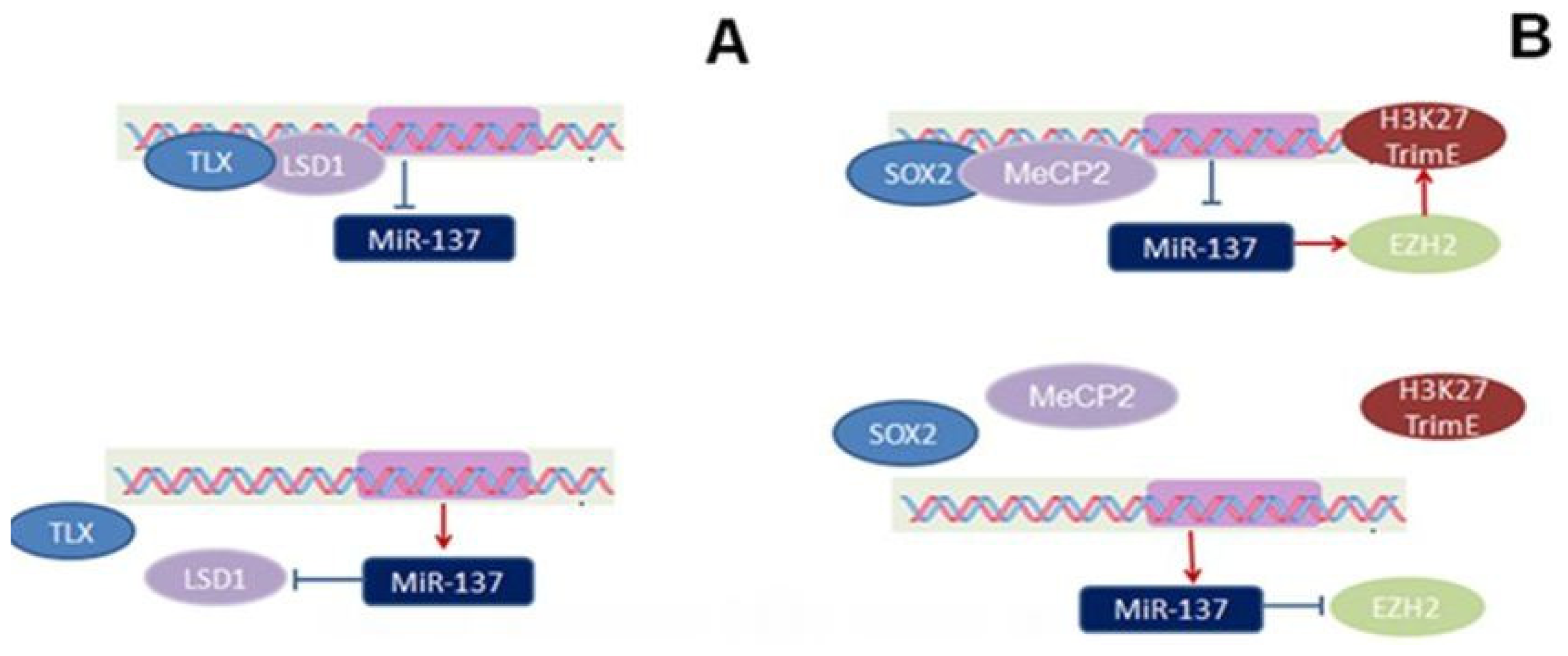
 , stimulatory effect found in embryos:
, stimulatory effect found in embryos:
 , Inhibitory effect found in adults:
, Inhibitory effect found in adults:
 , stimulatory effect found in adults:
, stimulatory effect found in adults:
 .
.
 , stimulatory effect found in embryos:
, stimulatory effect found in embryos:
 , Inhibitory effect found in adults:
, Inhibitory effect found in adults:
 , stimulatory effect found in adults:
, stimulatory effect found in adults:
 .
.
| Population | Number of participants | Genotype frequencies (%) | p-value | Correlated phenotypes | p-value | |||
|---|---|---|---|---|---|---|---|---|
| TT | GT | GG | ||||||
| American, Canadian (Lett et al.) | 510 | SZ | 67.45 | 29.8 | 2.75 | >0.05 * | age-at-onset and white matter integrity | 3.1 × 10−5 * |
| 121 | CTR | 62.81 | 33.06 | 4.13 | 3.88 × 10−8 * | |||
| Australian (Green et al.) | 491 | SZ | 64.97 | 31.16 | 3.87 | 0.85 * | cognitive deficits in combination with greater severity of negative symptoms | 0.017 * |
| 328 | CTR | 62.5 | 33.23 | 4.27 | ||||
| Scottish (Whalley et al.) | 44 | SZ | 59.09 | 38.64 | 2.27 | >0.05 | activation in the posterior right medial frontal gyrus, BA 6 | <0.001 |
| 81 | CTR | 61.73 | 35.8 | 2.47 | ||||
| American (Theo et al.) | 48 | SZ | 81.25 | 18.75 | 0 | 0.47 * | left DLPFC activation | 0.02 * |
| 63 | CTR | 73.02 | 23.81 | 3.17 | ||||
| Chinese (Guan et al.) | 1430 | SZ | 77.7 | 20.8 | 1.5 | 0.023 | ||
| 1570 | CTR | 74 | 23.6 | 2.4 | ||||
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yin, J.; Lin, J.; Luo, X.; Chen, Y.; Li, Z.; Ma, G.; Li, K. miR-137: A New Player in Schizophrenia. Int. J. Mol. Sci. 2014, 15, 3262-3271. https://doi.org/10.3390/ijms15023262
Yin J, Lin J, Luo X, Chen Y, Li Z, Ma G, Li K. miR-137: A New Player in Schizophrenia. International Journal of Molecular Sciences. 2014; 15(2):3262-3271. https://doi.org/10.3390/ijms15023262
Chicago/Turabian StyleYin, Jingwen, Juda Lin, Xudong Luo, Yanyan Chen, Zheng Li, Guoda Ma, and Keshen Li. 2014. "miR-137: A New Player in Schizophrenia" International Journal of Molecular Sciences 15, no. 2: 3262-3271. https://doi.org/10.3390/ijms15023262




