The Proteasome Activator PA28γ, a Negative Regulator of p53, Is Transcriptionally Up-Regulated by p53
Abstract
:1. Introduction
2. Results
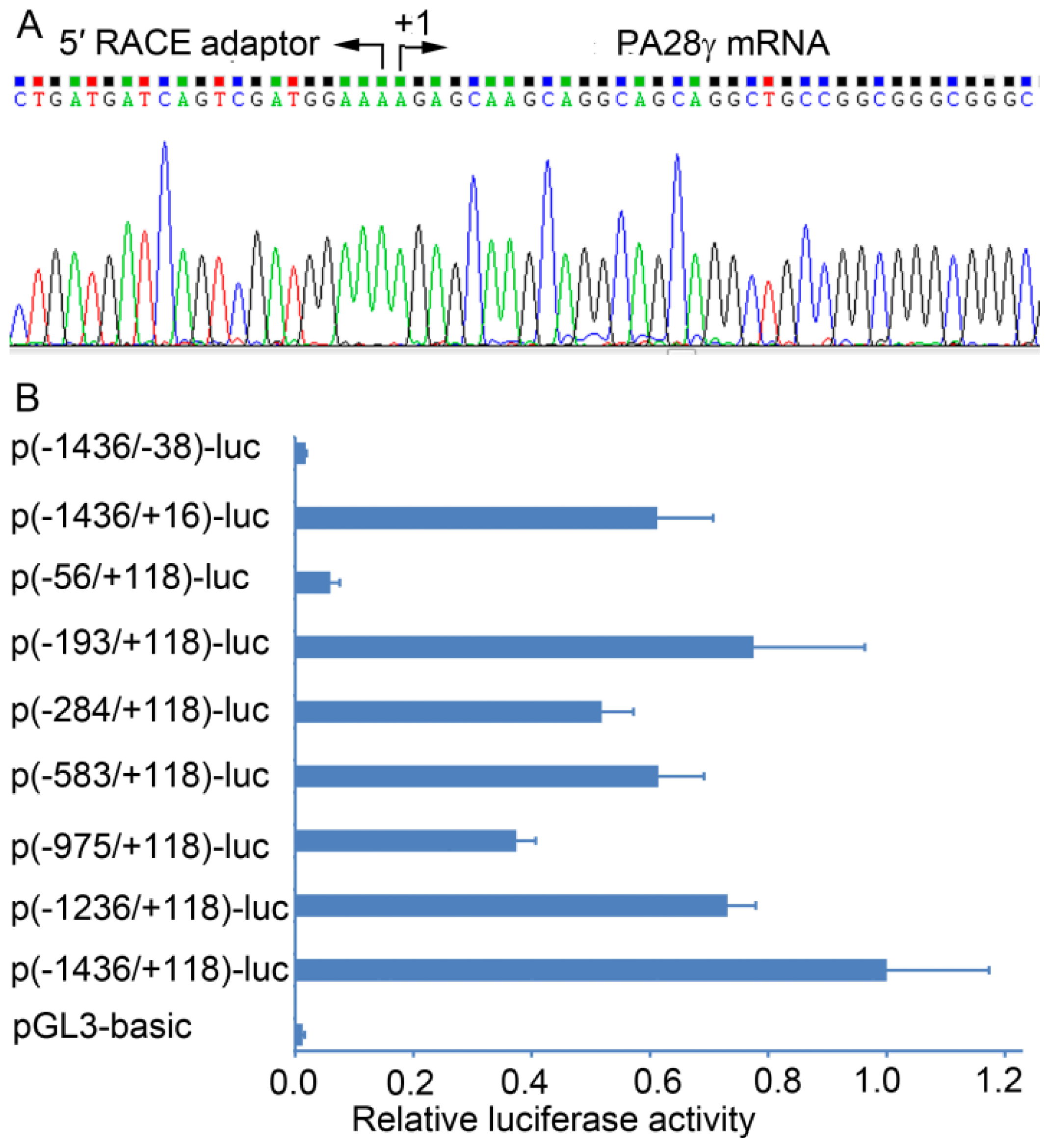
2.1. Determination of the Transcription Start Site (TSS) of the Human PA28γ Gene
2.2. Identification of the Basal Promoter of the Human PA28γ Gene
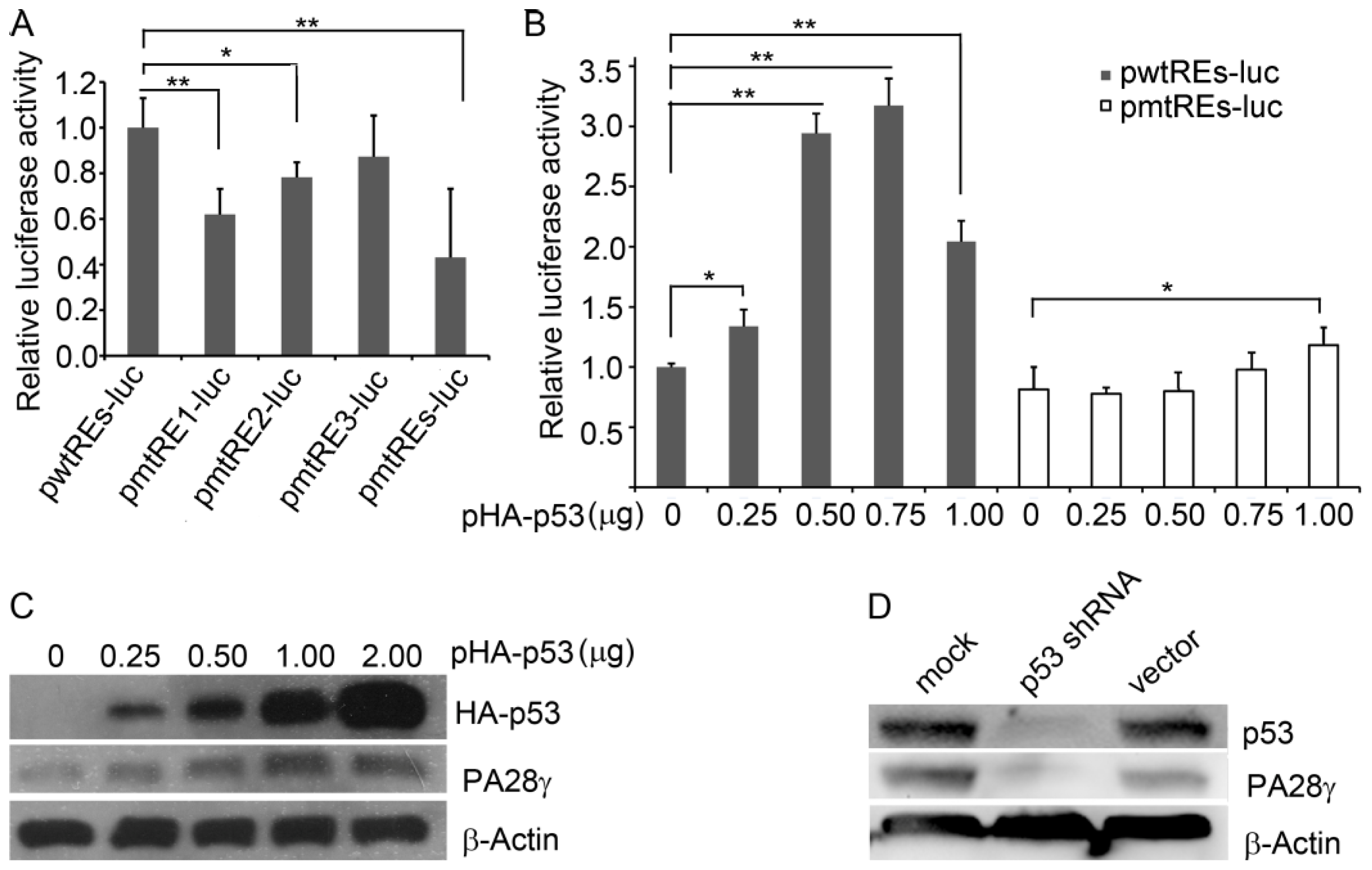
2.3. p53 Binds to the Promoter of the Human PA28γ Gene
2.4. p53 Stimulates the Transcription of Human PA28γ
2.5. p53-Induced Transcription of PA28γ Is Suppressed by PA28γ Itself
3. Discussion
4. Materials and Methods
4.1. Plasmid Constructs
4.2. Computational Analysis of the Promoter
4.3. Cell Culture and Transfection
4.4. Luciferase Assay
4.5. RLM-RACE Assay
4.6. ChIP Assay
4.7. Biotinylated DNA Affinity Precipitation
4.8. RNA Interference
4.9. Statistical Analysis
5. Conclusions
Acknowledgments
Conflicts of Interest
- Author ContributionsJ.-L.Z. conceived and designed the experiments. Z.-X.W., D.-M.Y., Y.-M.Z., X.Y., J.Z. and Z.-X.X. performed the experiments. Z.-X.W., D.-M.Y. and J.-L.Z. analyzed the data. J.-L.Z. wrote the manuscript. All authors read and approved the final manuscript.
References
- Tanaka, K. The proteasome: Overview of structure and functions. Proc. Jpn. Acad. Ser. B 2009, 85, 12–36. [Google Scholar]
- Mao, I.; Liu, J.; Li, X.; Luo, H. REGγ, a proteasome activator and beyond? Cell. Mol. Life Sci 2008, 65, 3971–3980. [Google Scholar]
- Yu, G.; Zhao, Y.; He, J.; Lonard, D.M.; Mao, C.A.; Wang, G.; Li, M.; Li, X. Comparative analysis of REGγ expression in mouse and human tissues. J. Mol. Cell Biol 2010, 2, 192–198. [Google Scholar]
- Murata, S.; Kawahara, H.; Tohma, S.; Yamamoto, K.; Kasahara, M.; Nabeshima, Y.; Tanaka, K.; Chiba, T. Growth retardation in mice lacking the proteasome activator PA28γ. J. Biol. Chem 1999, 274, 38211–38215. [Google Scholar]
- Chen, X.; Barton, L.F.; Chi, Y.; Clurman, B.E.; Roberts, J.M. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGγ proteasome. Mol. Cell 2007, 26, 843–852. [Google Scholar]
- Li, X.; Amazit, L.; Long, W.; Lonard, D.M.; Monaco, J.J.; O’Malley, B.W. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGγ-proteasome pathway. Mol. Cell 2007, 26, 831–842. [Google Scholar]
- Kobayashi, T.; Wang, J.; Al-Ahmadie, H.; Abate-Shen, C. ARF regulates the stability of p16 protein via REGγ-dependent proteasome degradation. Mol. Cancer Res 2013, 11, 828–833. [Google Scholar]
- Zhang, Z.; Zhang, R. Proteasome activator PA28γ regulates p53 by enhancing its MDM2-mediated degradation. EMBO J 2008, 27, 852–864. [Google Scholar]
- Liu, J.; Yu, G.; Zhao, Y.; Zhao, D.; Wang, Y.; Wang, L.; Liu, J.; Li, L.; Zeng, Y.; Dang, Y.; et al. REGγ modulates p53 activity by regulating its cellular localization. J. Cell Sci 2010, 123, 4076–4084. [Google Scholar]
- Roessler, M.; Rollinger, W.; Mantovani-Endl, L.; Hagmann, M.L.; Palme, S.; Berndt, P.; Engel, A.M.; Pfeffer, M.; Karl, J.; Bodenmüller, H.; et al. Identification of PSME3 as a novel serum tumor marker for colorectal cancer by combining two-dimensional polyacrylamide gel electrophoresis with a strictly mass spectrometry-based approach for data analysis. Mol. Cell Proteomics 2006, 5, 2092–2101. [Google Scholar]
- Okamura, T.; Taniguchi, S.; Ohkura, T.; Yoshida, A.; Shimizu, H.; Sakai, M.; Maeta, H.; Fukui, H.; Ueta, Y.; Hisatome, I.; et al. Abnormally high expression of proteasome activator-γ in thyroid neoplasm. J. Clin. Endocrinol. Metab 2003, 88, 1374–1383. [Google Scholar]
- Wang, X.; Tu, S.; Tan, J.; Tian, T.; Ran, L.; Rodier, J.F.; Ren, G. REGγ: A potential marker in breast cancer and effect on cell cycle and proliferation of breast cancer cell. Med. Oncol 2011, 28, 31–41. [Google Scholar]
- Li, L.P.; Cheng, W.B.; Li, H.; Li, W.; Yang, H.; Wen, D.H.; Tang, Y.D. Expression of proteasome activator REGγ in human laryngeal carcinoma and associations with tumor suppressor proteins. Asian Pac. J. Cancer Prev 2012, 13, 2699–2703. [Google Scholar]
- Li, X.; Lonard, D.M.; Jung, S.Y.; Malovannaya, A.; Feng, Q.; Qin, J.; Tsai, S.Y.; Tsai, M.J.; O’Malley, B.W. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGγ proteasome. Cell 2006, 124, 381–392. [Google Scholar]
- Moriishi1, K.; Okabayashi1, T.; Nakai1, K.; Moriya, K.; Koike, K.; Murata, S.; Chiba, T.; Tanaka, K.; Suzuki, E.; Suzuki, T.; et al. Proteasome activator PA28γ-dependent nuclear retention and degradation of hepatitis C virus core protein. J. Virol 2003, 77, 10237–10249. [Google Scholar]
- Ying, H.; Furuya, F.; Zhao, L.; Araki, O.; West, B.L.; Hanover, J.A.; Willingham, M.C.; Cheng, S.Y. Aberrant accumulation of PTTG1 induced by a mutated thyroid hormone β receptor inhibits mitotic progression. J. Clin. Invest 2006, 116, 2972–2984. [Google Scholar]
- Levy-Barda, A.; Lerenthal, Y.; Davis, A.J.; Chung, Y.M.; Essers, J.; Shao, Z.; van Vliet, N.; Chen, D.J.; Hu, M.C.T.; Kanaar, R.; et al. Involvement of the nuclear proteasome activator PA28γ in the cellular response to DNA double-strand breaks. Cell Cycle 2011, 10, 4300–4310. [Google Scholar]
- Zannini, L.; Lecis, D.; Buscemi, G.; Carlessi, L.; Gasparini, P.; Fontanella, E.; Lisanti, S.; Barton, L.; Delia, D. REGγ proteasome activator is involved in the maintenance of chromosomal stability. Cell Cycle 2008, 7, 504–512. [Google Scholar]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol 2006, 7, S10. [Google Scholar]
- Solovyev, V.; Salamov, A. The Gene-Finder computer tools for analysis of human and model organisms genome sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol 1997, 5, 294–302. [Google Scholar]
- Reese, M.G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem 2001, 26, 51–56. [Google Scholar]
- Ohler, U.; Stemmer, G.; Harbeck, S.; Niemann, H. Stochastic segment models of eukaryotic promoter regions. Pac. Symp. Biocomput 2000, 5, 377–388. [Google Scholar]
- Maruyama, K.; Sugano, S. Oligo-capping: A simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 1994, 138, 171–174. [Google Scholar]
- Broos, S.; Hulpiau, P.; Galle, J.; Hooghe, B.; van Roy, F.; de Bleser, P. ConTra v2: A tool to identify transcription factor binding sites across species, update 2011. Nucleic Acids Res 2011, 39, W74–W78. [Google Scholar]
- Cartharius, K.; Frech, K.; Grote, K.; Klocke, B.; Haltmeier, M.; Klingenhoff, A.; Frisch, M.; Bayerlein, M.; Werner, T. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics 2005, 21, 2933–2942. [Google Scholar]
- Wu, X.; Bayle, J.H.; Olson, D.; Levine, A.J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev 1993, 7, 1126–1132. [Google Scholar]
- Zhou, J.; Qiao, X.; Xiao, L.; Sun, W.; Wang, L.; Li, H.; Wu, Y.; Ding, X.; Hu, X.; Zhou, C.; et al. Identification and characterization of the novel protein CCDC106 that interacts with p53 and promotes its degradation. FEBS Lett 2010, 584, 1085–1090. [Google Scholar]
- Zhang, H.; Chen, H.; Luo, H.; An, J.; Sun, L.; Mei, L.; He, C.; Jiang, L.; Jiang, W.; Xia, K.; et al. Functional analysis of Waardenburg syndrome-associated PAX3 and SOX10 mutations: Report of a dominant-negative SOX10 mutation in Waardenburg syndrome type II. Hum. Genet 2012, 131, 491–503. [Google Scholar]


© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wan, Z.-X.; Yuan, D.-M.; Zhuo, Y.-M.; Yi, X.; Zhou, J.; Xu, Z.-X.; Zhou, J.-L. The Proteasome Activator PA28γ, a Negative Regulator of p53, Is Transcriptionally Up-Regulated by p53. Int. J. Mol. Sci. 2014, 15, 2573-2584. https://doi.org/10.3390/ijms15022573
Wan Z-X, Yuan D-M, Zhuo Y-M, Yi X, Zhou J, Xu Z-X, Zhou J-L. The Proteasome Activator PA28γ, a Negative Regulator of p53, Is Transcriptionally Up-Regulated by p53. International Journal of Molecular Sciences. 2014; 15(2):2573-2584. https://doi.org/10.3390/ijms15022573
Chicago/Turabian StyleWan, Zhen-Xing, Dong-Mei Yuan, Yi-Ming Zhuo, Xin Yi, Ji Zhou, Zao-Xu Xu, and Jian-Lin Zhou. 2014. "The Proteasome Activator PA28γ, a Negative Regulator of p53, Is Transcriptionally Up-Regulated by p53" International Journal of Molecular Sciences 15, no. 2: 2573-2584. https://doi.org/10.3390/ijms15022573



