Isolation and Molecular Characterization of Biofouling Bacteria and Profiling of Quorum Sensing Signal Molecules from Membrane Bioreactor Activated Sludge
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation and Screening of AHLs Producing Bacteria
2.2. Biofilm Formation among AHLs Producing Bacterial Isolates
2.3. Identification of AHLs Producing Bacteria
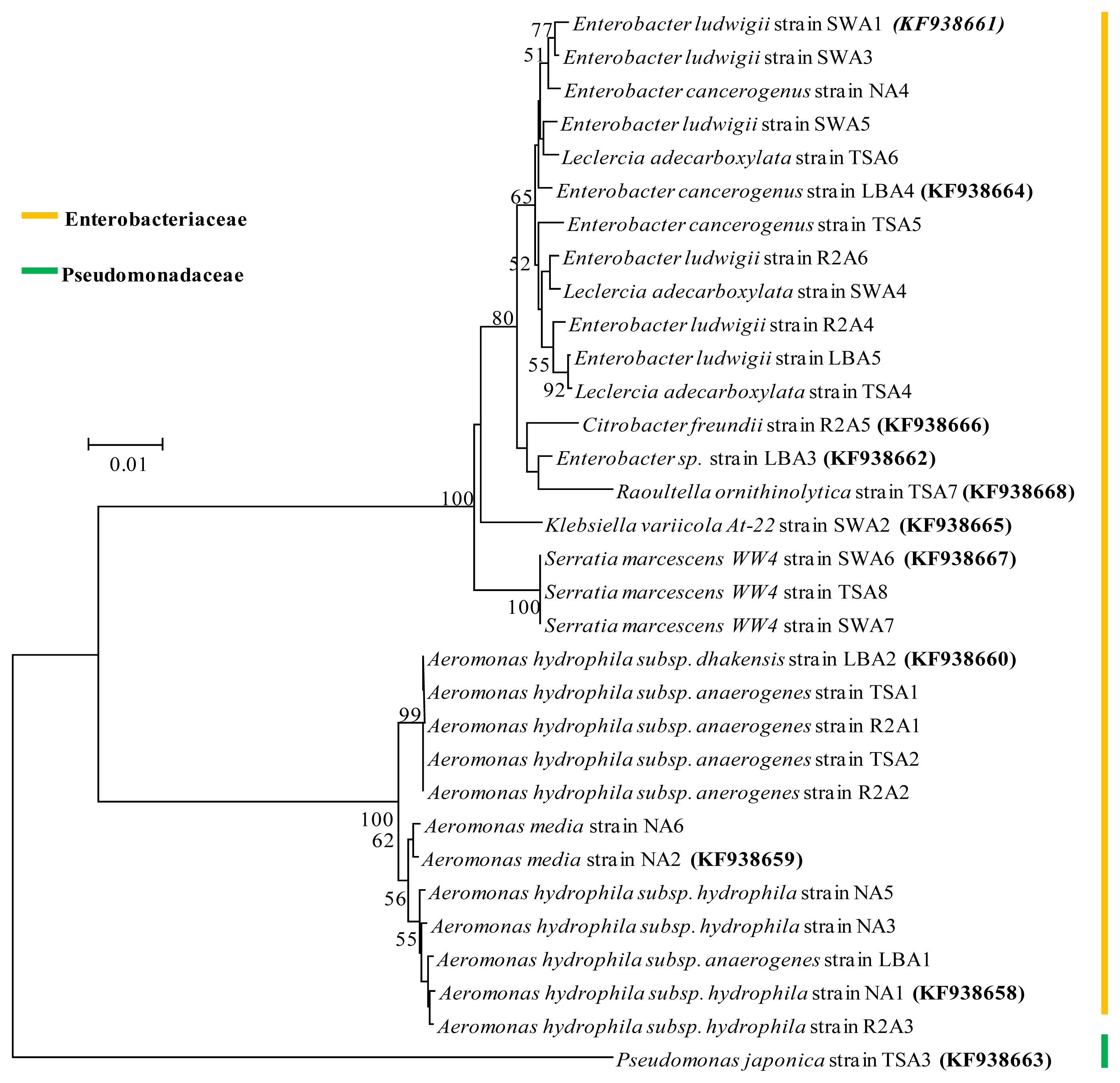
2.4. Phylogenetic Analysis
2.5. Nucleotide Sequence Accession Numbers
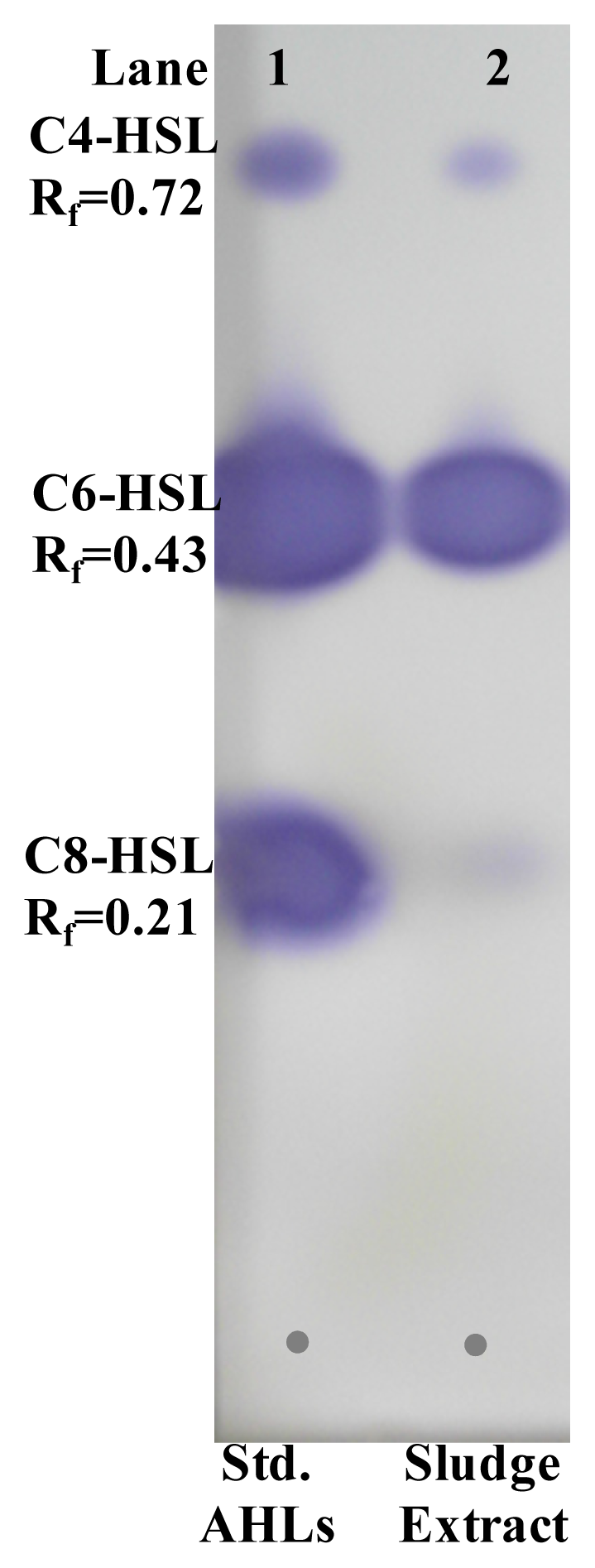
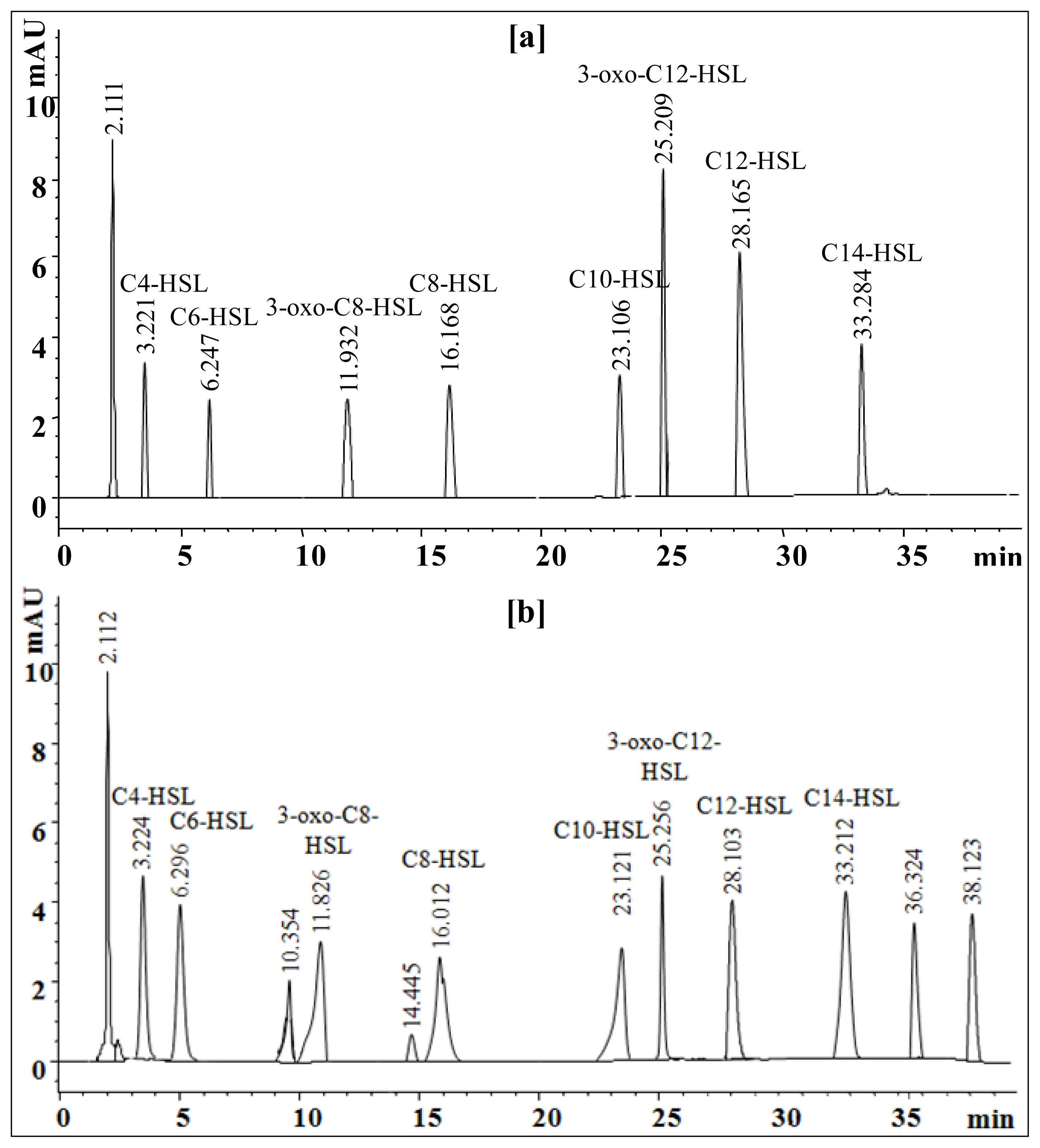
2.6. Extraction and Characterization of Sludge AHLs
3. Experimental Section
3.1. Chemicals and Microbiological Media
3.2. Standard N-acyl Homoserine Lactones
3.3. Quorum Sensing Reporter’s Strains
3.4. Collection of Activated Sludge
3.5. Isolation of Bacteria
3.6. Screening of Bacterial Isolates for AHLs Production
3.7. Microtiter Plate Biofilm Formation Assay
3.8. Identification of AHLs Producing Bacteria
3.9. Phylogenetic Analysis of AHLs Producing Bacteria Strains
3.10. Nucleotide Sequence Accession Number
3.11. Extraction of AHLs from Activated Sludge
3.12. Detection of Crude AHLs by Agar Plate Cross-Feeding Bioassay
3.13. Thin Layer Chromatography (TLC) for AHL Identification
3.14. High Performance Liquid Chromatography (HPLC) Analysis of AHLs
3.15. Statistical Analysis
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Drews, A. Membrane fouling in membrane bioreactors—Characterisation, contradictions, cause and cures. J. Membr. Sci 2010, 363, 1–28. [Google Scholar]
- Lee, J.W.; Jutidamrongphan, W.; Park, K.Y.; Moon, S.; Park, C. Advanced treatment of wastewater from food waste disposer in modified Ludzack-Ettinger type membrane bioreactor. Environ. Eng. Res 2012, 17, 59–63. [Google Scholar]
- Miura, Y.; Watanabe, Y.; Okabe, S. Membrane biofouling in pilotscale membrane bioreactors (MBRs) treating municipal wastewater: Impact of biofilm formation. Environ. Sci. Technol 2007, 41, 632–638. [Google Scholar]
- Lim, S.Y.; Kim, S.; Yeon, K.M.; Sang, B.I.; Chun, J.; Lee, C.H. Correlation between microbial community structure and biofouling in a laboratory scale membrane bioreactor with synthetic wastewater. Desalination 2012, 287, 209–215. [Google Scholar]
- Wagner, M.; Loy, A. Bacterial community composition and function in sewage treatment systems. Curr. Opin. Microbiol 2002, 13, 218–227. [Google Scholar]
- Chong, G.; Kimyon, O.; Rice, S.A.; Kjelleberg, S.; Manefield, M. The presence and role of bacterial quorum sensing in activated sludge. Microb. Biotechnol 2012, 5, 621–633. [Google Scholar]
- Yeon, K.M.; Cheong, W.S.; Oh, H.S.; Lee, W.N.; Hwang, B.K.; Lee, C.H.; Beyenal, H.; Lewandowski, Z. Quorum sensing: A new biofouling control paradigm in a membrane bioreactor for advanced wastewater treatment. Environ. Sci. Technol 2009, 43, 380–385. [Google Scholar]
- Chen, J.W.; Koh, C.L.; Sam, C.K.; Yin, W.F.; Chan, K.G. Short chain N-acyl homoserine lactone production by soil isolate Burkholderia sp. strain A9. Sensors 2013, 13, 13217–13227. [Google Scholar]
- Hugenholtz, P.; Goebel, B.M.; Pace, N.R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol 1998, 180, 4765–4774. [Google Scholar]
- Alex, A.; Silva, V.; Vasconcelos, V.; Antunes, A. Evidence of unique and generalist microbes in distantly related sympatric intertidal marine sponges (Porifera: Demospongiae). PLoS One 2013, 8, e80653. [Google Scholar]
- Johnson, L.R. Microcolony and biofilm formation as a survival strategy for bacteria. J. Theor. Biol 2008, 251, 24–34. [Google Scholar]
- Rinaudi, L.V.; Giordano, W. An integrated view of biofilm formation in rhizobia. FEMS Microbiol. Lett 2010, 304, 1–11. [Google Scholar]
- Dong, Y.H.; Zhang, X.F.; An, S.W.; Xu, J.L.; Zhang, L.H. A novel two-component system BqsS-BqsR modulates quorum sensing-dependent biofilm decay in Pseudomonas aeruginosa. Commun. Integr. Biol 2008, 1, 88–96. [Google Scholar]
- Rasmussen, T.B.; Bjarnsholt, T.; Skindersoe, M.E.; Hentzer, M.; Kristoffersen, P.; Kote, M.; Nielsen, J.; Eberl, L.; Givskov, M. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol 2005, 187, 1799–1814. [Google Scholar]
- Daniels, R.; Vanderleyden, J.; Michiels, J. Quorum sensing and swarming migration in bacteria. FEMS Microbiol. Rev 2004, 28, 261–289. [Google Scholar]
- Stoodley, P.; Wilson, S.; Hall-Stoodley, L.; Boyle, J.D.; Lappin-Scott, H.M.; Costerton, J.W. Growth and detachment of cell clusters from mature mixed-species biofilms. Appl. Environ. Microbiol 2001, 67, 5608–5613. [Google Scholar]
- Adegboye, M.F.; Babalola, O.O. Phylogenetic characterization of culturable antibiotic producing Streptomyces from rhizospheric soils. Mol. Biol 2013, S1. [Google Scholar] [CrossRef]
- Wang, H.; Cai, T.; Weng, M.; Zhou, J.; Cao, H.; Zhong, Z.; Zhu, J. Conditional production of acyl-homoserine lactone-type quorum-sensing signals in clinical isolates of enterobacteria. J. Med. Microbiol 2006, 55, 1751–1753. [Google Scholar]
- McClean, K.H.; Winson, M.K.; Fish, L.; Taylor, A.; Chhabra, S.R. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 1997, 143, 3703–3711. [Google Scholar]
- Ravn, L.; Christensen, A.B.; Molin, S.; Givskov, M.; Gram, L. Methods for detecting acylated homoserine lactones produced by Gram-negative bacteria and their application in studies of AHL-production kinetics. J. Microbiol. Methods 2001, 44, 239–251. [Google Scholar]
- Kim, S.R.; Oh, H.S.; Jo, S.J.; Yeon, K.M.; Lee, C.H.; Lim, D.J.; Lee, C.H.; Lee, J.K. Biofouling control with bead-entrapped quorum quenching bacteria in membrane bioreactors: Physical and biological effects. Environ. Sci. Technol 2013, 47, 836–842. [Google Scholar]
- Anbazhagan, D.; Mansor, M.; Yan, G.O.S.; Yusof, M.Y.M.; Hassan, H.; Sekaran, S.D. Detection of quorum sensing signal molecules and identification of an autoinducer synthase gene among biofilm forming clinical isolates of Acinetobacter spp. PLoS One 2012, 7, e36696. [Google Scholar]
- O’Toole, G.A.; Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis. Mol. Microbiol 1998, 8, 449–461. [Google Scholar]
- Biswa, P.; Doble, M. Production of acylated homoserine lactone by Gram-positive bacteria isolated from marine water. FEMS Microbiol. Lett 2013, 343, 34–41. [Google Scholar]
- Ponnusamy, K.; Paul, D.; Kweon, J. Inhibition of quorum sensing mechanism and Aeromonas hydrophila biofilm formation by vanillin. Env. Eng. Sci 2009, 26, 1359–1363. [Google Scholar]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol 2000, 7, 203–214. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W-improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 1994, 22, 4673–4680. [Google Scholar]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol 2007, 24, 1596–1599. [Google Scholar]
- Oh, H.S.; Yeon, K.M.; Yang, C.S.; Kim, S.R.; Lee, C.H.; Park, S.Y.; Han, J.Y.; Lee, J.K. Control of membrane biofouling in MBR for wastewater treatment by quorum quenching bacteria encapsulated in microporous membrane. Environ. Sci. Technol 2012, 46, 4877–4884. [Google Scholar]
- Kumari, A.; Pasini, P.; Daunert, S. Detection of bacterial quorum sensing N-acyl homoserine lactones in clinical samples. Anal. Bioanal. Chem 2008, 391, 1619–1627. [Google Scholar]
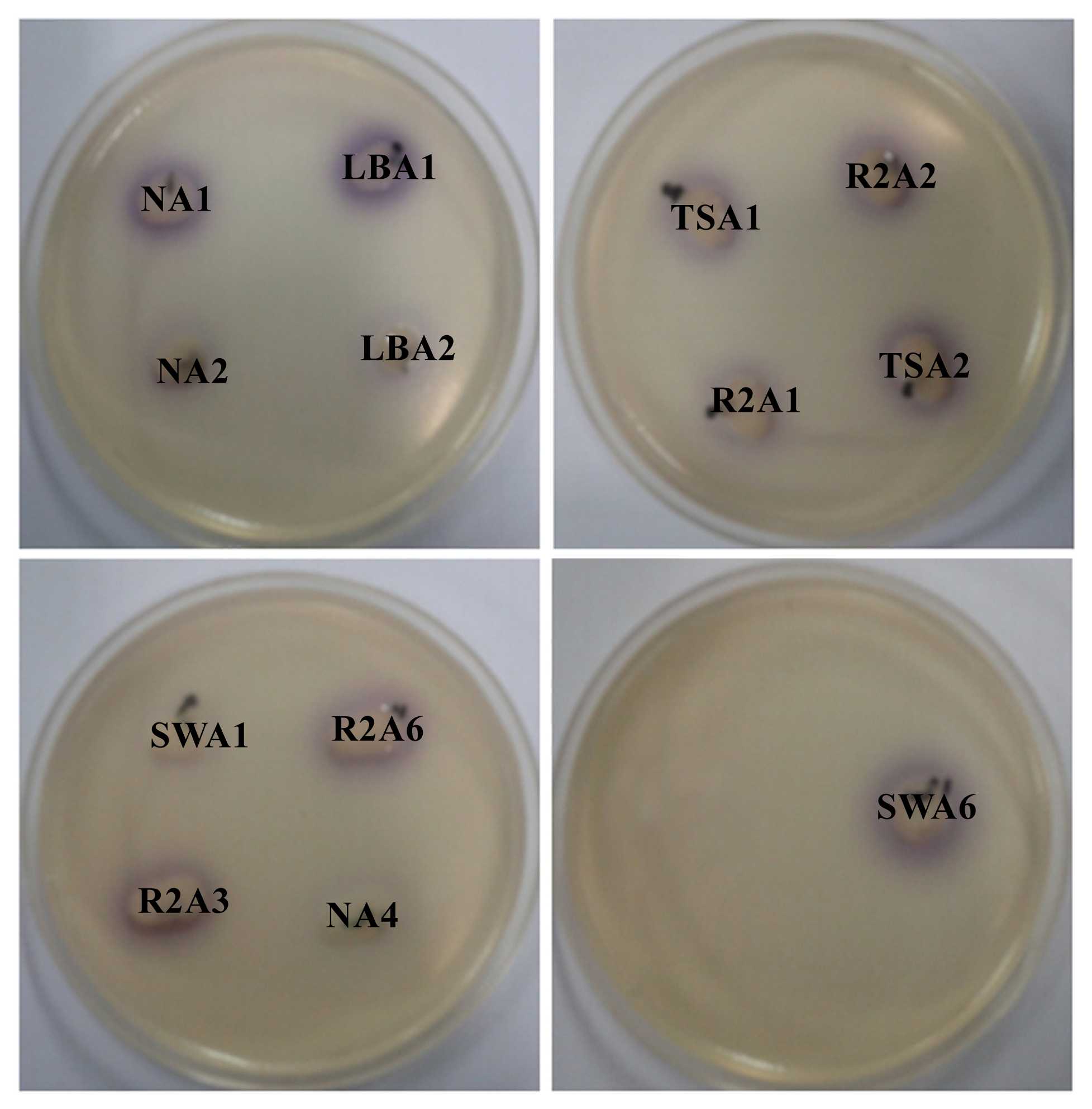
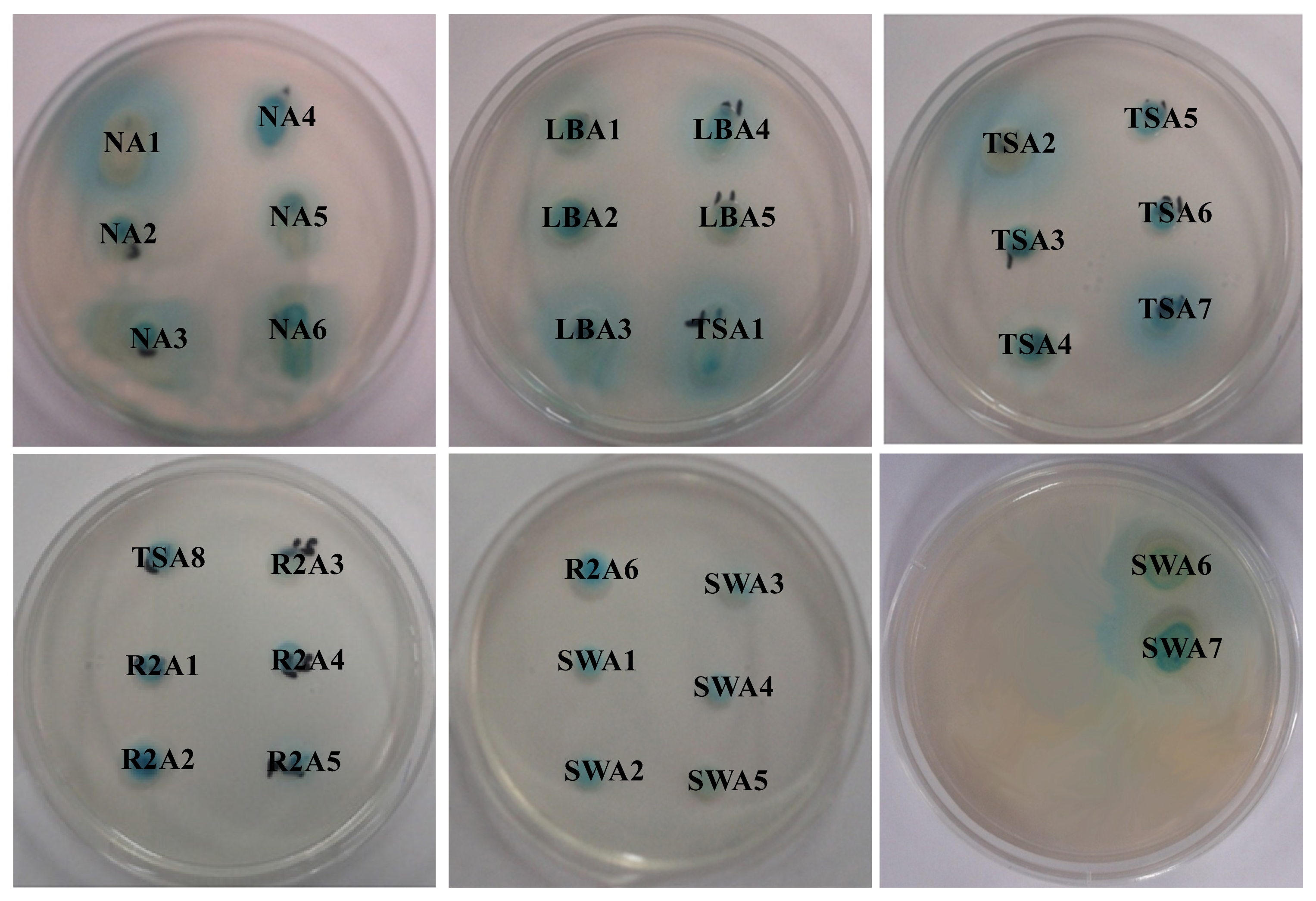


| Isolate numbers | Isolate identifier | Reporter strains | |
|---|---|---|---|
| C. violaceum CV026 | A. tumefaciens A136 | ||
| 1 | NA1 | +++ | +++ |
| 2 | NA2 | ++ | +++ |
| 3 | NA3 | -- | ++ |
| 4 | NA4 | ++ | +++ |
| 5 | NA5 | -- | ++ |
| 6 | NA6 | -- | ++ |
| 7 | LBA1 | +++ | +++ |
| 8 | LBA2 | ++ | +++ |
| 9 | LBA3 | -- | +++ |
| 10 | LBA4 | -- | +++ |
| 11 | LBA5 | -- | ++ |
| 12 | TSA1 | +++ | +++ |
| 13 | TSA2 | +++ | +++ |
| 14 | TSA3 | -- | +++ |
| 15 | TSA4 | -- | ++ |
| 16 | TSA5 | -- | ++ |
| 17 | TSA6 | -- | ++ |
| 18 | TSA7 | -- | ++ |
| 19 | TSA8 | -- | +++ |
| 20 | R2A1 | +++ | +++ |
| 21 | R2A2 | +++ | +++ |
| 22 | R2A3 | +++ | ++ |
| 23 | R2A4 | -- | ++ |
| 24 | R2A5 | -- | ++ |
| 25 | R2A6 | +++ | +++ |
| 26 | SWA1 | ++ | +++ |
| 27 | SWA2 | -- | +++ |
| 28 | SWA3 | -- | ++ |
| 29 | SWA4 | -- | +++ |
| 30 | SWA5 | -- | +++ |
| 31 | SWA6 | ++ | ++ |
| 32 | SWA7 | -- | ++ |
| Isolate identifier | Sequence length (bp) | Max score | Identity (%) | Closest relative strain at database and its description | Accession number |
|---|---|---|---|---|---|
| NA1 | 1469 | 2678 | 99 | Aeromonas hydrophila subsp. hydrophila ATCC 7966 strain ATCC 7966 16S rRNA, complete sequence | NR_074841.1 |
| NA2 | 1470 | 2715 | 100 | Aeromonas media strain RM 16S rRNA, partial sequence | NR_036911.2 |
| NA3 | 1465 | 2684 | 100 | Aeromonas hydrophila subsp. hydrophila ATCC 7966 strain ATCC 7966 16S rRNA, complete sequence | NR_074841.1 |
| NA4 | 1464 | 2575 | 99 | Enterobacter cancerogenus strain LMG 2693 16S rRNA, partial sequence | NR_044977.1 |
| NA5 | 1474 | 2712 | 100 | Aeromonas hydrophila subsp. hydrophila ATCC 7966 strain ATCC 7966 16S rRNA, complete sequence | NR_074841.1 |
| NA6 | 1465 | 2684 | 100 | Aeromonas media strain RM 16S rRNA, partial sequence | NR_036911.2 |
| LBA1 | 1461 | 2641 | 100 | Aeromonas hydrophila subsp. anaerogenes strain CECT 4221 16S rRNA, partial sequence | NR_104824.1 |
| LBA2 | 1474 | 2684 | 99 | Aeromonas hydrophila subsp. dhakensis strain P21 16S rRNA, partial sequence | NR_042155.1 |
| LBA3 | 1396 | 2518 | 100 | Enterobacter sp. 638 strain 638 16S rRNA, complete sequence | NR_074777.1 |
| LBA4 | 1383 | 2505 | 100 | Enterobacter cancerogenus strain LMG 2693 16S rRNA, partial sequence | NR_044977.1 |
| LBA5 | 1459 | 2586 | 99 | Enterobacter ludwigii strain EN-119 16S rRNA, complete sequence | NR_042349.1 |
| TSA1 | 1471 | 2717 | 100 | Aeromonas hydrophila subsp. anaerogenes strain CECT 4221 16S rRNA, partial sequence | NR_104824.1 |
| TSA2 | 1476 | 2726 | 100 | Aeromonas hydrophila subsp. anaerogenes strain CECT 4221 16S rRNA, partial sequence | NR_104824.1 |
| TSA3 | 1460 | 2601 | 100 | Pseudomonas japonica strain IAM 15071 16S rRNA, partial sequence | NR_040992.1 |
| TSA4 | 1459 | 2623 | 100 | Leclercia adecarboxylata strain CIP 82.92 16S rRNA, complete sequence | NR_104933.1 |
| TSA5 | 1462 | 2647 | 99 | Enterobacter cancerogenus strain LMG 2693 16S rRNA, partial sequence | NR_044977.1 |
| TSA6 | 1462 | 2558 | 100 | Leclercia adecarboxylata strain CIP 82.92 16S rRNA, complete sequence | NR_104933.1 |
| TSA7 | 1461 | 2671 | 100 | Raoultella ornithinolytica B6 strain B6 16S rRNA, complete sequence | NR_102983.1 |
| TSA8 | 1463 | 2680 | 100 | Serratia marcescens WW4 strain WW4 16S rRNA, complete sequence | NR_102509.1 |
| R2A1 | 1475 | 2719 | 100 | Aeromonas hydrophila subsp. anaerogenes strain CECT 4221 16S rRNA, partial sequence | NR_104824.1 |
| R2A2 | 1465 | 2706 | 100 | Aeromonas hydrophila subsp. anaerogenes strain CECT 4221 16S rRNA, partial sequence | NR_104824.1 |
| R2A3 | 1476 | 2699 | 99 | Aeromonas hydrophila subsp. hydrophila ATCC 7966 strain ATCC 7966 16S rRNA, complete sequence | NR_074841.1 |
| R2A4 | 1455 | 2599 | 99 | Enterobacter ludwigii strain EN-119 16S rRNA, complete sequence | NR_042349.1 |
| R2A5 | 1465 | 2686 | 99 | Citrobacter freundii strain DSM 30039 16S rRNA, partial sequence | NR_028894.1 |
| R2A6 | 1464 | 2638 | 99 | Enterobacter ludwigii strain EN-119 16S rRNA, complete sequence | NR_042349.1 |
| SWA1 | 1490 | 2571 | 98 | Enterobacter ludwigii strain EN-119 16S rRNA, complete sequence | NR_042349.1 |
| SWA2 | 1460 | 2676 | 99 | Klebsiella variicola At-22 strain At-22 16S rRNA, complete sequence | NR_074729.1 |
| SWA3 | 1450 | 2555 | 100 | Enterobacter ludwigii strain EN-119 16S rRNA, complete sequence | NR_042349.1 |
| SWA4 | 1462 | 2619 | 99 | Leclercia adecarboxylata strain CIP 82.92 16S rRNA, complete sequence | NR_104933.1 |
| SWA5 | 1458 | 2619 | 99 | Enterobacter ludwigii strain EN-119 16S rRNA, complete sequence | NR_042349.1 |
| SWA6 | 1462 | 2673 | 100 | Serratia marcescens WW4 strain WW4 16S rRNA, complete sequence | NR_102509.1 |
| SWA7 | 1463 | 2673 | 99 | Serratia marcescens WW4 strain WW4 16S rRNA, complete sequence | NR_102509.1 |
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lade, H.; Paul, D.; Kweon, J.H. Isolation and Molecular Characterization of Biofouling Bacteria and Profiling of Quorum Sensing Signal Molecules from Membrane Bioreactor Activated Sludge. Int. J. Mol. Sci. 2014, 15, 2255-2273. https://doi.org/10.3390/ijms15022255
Lade H, Paul D, Kweon JH. Isolation and Molecular Characterization of Biofouling Bacteria and Profiling of Quorum Sensing Signal Molecules from Membrane Bioreactor Activated Sludge. International Journal of Molecular Sciences. 2014; 15(2):2255-2273. https://doi.org/10.3390/ijms15022255
Chicago/Turabian StyleLade, Harshad, Diby Paul, and Ji Hyang Kweon. 2014. "Isolation and Molecular Characterization of Biofouling Bacteria and Profiling of Quorum Sensing Signal Molecules from Membrane Bioreactor Activated Sludge" International Journal of Molecular Sciences 15, no. 2: 2255-2273. https://doi.org/10.3390/ijms15022255







