Identification of Protein Markers in Patients Infected with Plasmodium knowlesi, Plasmodium falciparum and Plasmodium vivax
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Candidate Biomarkers by iTRAQ
2.2. Analysis of ASKc-Depleted Sera by iTRAQ

| Protein | M1/C1 | M2/C1 | M3/C1 |
|---|---|---|---|
| HAP | 0.540 | 0.633 | 0.435 |
| CRP | 3.243 | 2.154 | 1.925 |
| CADM4 | 6.386 | 3.688 | 9.760 |
2.3. Validation of HAP by Protein Expression
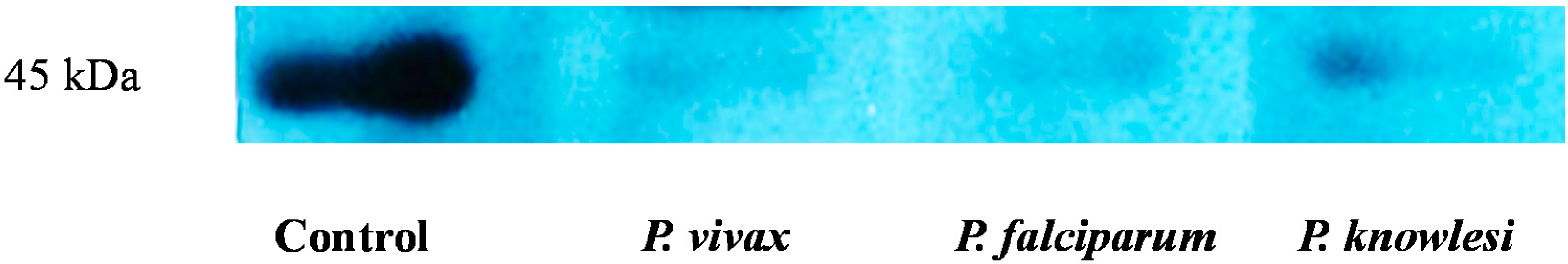
3. Experimental Section
3.1. Sample Collection
3.2. Albumin Depletion
3.3. Sample Preparation and Labeling for iTRAQ
3.4. Liquid Chromatography-Tandem MS (LC-MS/MS) Analysis
3.5. Data Analysis
3.6. Western Blotting
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- World Malaria Report. 2013. Available online: http://www.who.int/malaria/publication/world_malaria_report/en/ (accessed on 6 October 2014).
- Tuteja, R. Malaria—An overview. FEBS J. 2007, 274, 4670–4679. [Google Scholar] [CrossRef]
- Daneshvar, C.; Davis, T.M.E.; Cox-Singh, J.; Rafa’ee, M.Z.; Zakaria, S.K.; Divis, P.C.S.; Singh, B. Clinical and laboratory features of human Plasmodium knowlesi infection. Clin. Infect. Dis. 2009, 49, 852–860. [Google Scholar] [CrossRef]
- Adam, I.; Omer, E.S.M.; Salih, A.; Malik, A.H.K.E.M. Perceptions of the causes of malaria and of its complications, treatment and prevention among midvies and pregnant women of Eastern Sudan. J. Public Health 2008, 16, 129–132. [Google Scholar] [CrossRef]
- Jongwutiwes, S.; Putaporntip, C.; Iwasaki, T.; Sata, T.; Kanbara, H. Naturally acquired Plasmodium knowlesi malaria in human, Thailand. Emerg. Infect. Dis. 2004, 10, 2211–2213. [Google Scholar] [CrossRef]
- Conroy, A.L.; Lafferty, E.I.; Lovegrove, F.E.; Krudsood, S.; Tangpukdee, N.; Liles, W.C.; Kain, K.C. Whole blood angiopoietin-1 and -2 levels discriminate cerebral and severe (non-cerebral) malaria from uncomplicated malaria. Malar. J. 2009, 8. [Google Scholar] [CrossRef]
- Armah, H.B.; Wilson, N.O.; Sarfo, B.Y.; Powell, M.D.; Bond, V.C.; Anderson, W.; Adjei, A.A.; Gyasi, R.K.; Tettey, Y.; Wiredu, E.K.; et al. Cerebrospinal fluid and serum biomarkers of cerebral malaria mortality in Ghanaian children. Malar. J. 2007, 6. [Google Scholar] [CrossRef]
- Sohail, M.; Kaul, A.; Raziuddin, M.; Adak, T. Decreased glutathione-S-transferase activity: Diagnostic and protective role in vivax malaria. Clin. Biochem. 2007, 40, 377–382. [Google Scholar] [CrossRef]
- Chattopadhyay, R.; Velmurugan, S.; Chakiath, C.; Donkor, L.A.; Milhous, W.; Barnwell, J.W.; Collins, W.E.; Hoffman, S.L. Establishment of an In vitro assay for assessing the liver stages of Plasmodiun. vivax malaria. PLoS One 2010, 5, e14275. [Google Scholar] [CrossRef]
- Nirmalan, N.; Sims, P.F.G.; Hyde, J.E. Quantitative proteomics of the human malaria parasite Plasmodium falciparum and its application to studies of development and inhibition. Mol. Microbiol. 2004, 52, 1187–1199. [Google Scholar] [CrossRef]
- Mu, A.K.W.; Lim, B.K.; Hashim, O.H.; Shuib, A.S. Detection of differential levels of proteins in the urine of patients with endometrial cancer: Analysis using two-dimensional gel electrophoresis and O-glycan binding lectin. Int. J. Mol. Sci. 2012, 13, 9489–9501. [Google Scholar] [CrossRef]
- Mu, A.K.W.; Lim, B.K.; Hashim, O.H.; Shuib, A.S. Identification of O-glycosylated proteins that are aberrantly excreted in the urine of patients with early stage ovarian cancer. Int. J. Mol. Sci. 2013, 14, 7923–7931. [Google Scholar] [CrossRef]
- Mu, A.K.W.; Chan, Y.S.; Kang, S.S.; Azman, S.N.; Zain, R.B.; Chai, W.L.; Chen, Y. Detection of host immunogenic proteins in the saliva of patients with oral squamous cell carcinoma. J. Immunoass. Immunochem. 2014, 35, 183–193. [Google Scholar] [CrossRef]
- Meyfour, A.; Tavirani, M.R.; Sadeghi, M.R. Common proteomic technologies, applications and their limitations. J. Paramed. Sci. 2008, 4, 115–125. [Google Scholar]
- Schuchard, M.D.; Melm, C.D.; Crawford, A.S.; Chapman, H.A.; Cockrill, S.L.; Ray, K.B.; Mehigh, R.J.; Kappel, W.K.; Scott, G.B.I. Novel immunoaffinity depletion technology with increased binding capacity removes approximately 97% of high abundance human plasma proteins. In Prceedings of the ASMS Conference, San Antonio, TX, USA, 5–9 June 2005.
- Ray, S.; Renu, D.; Srivastava, R.; Gdlapalli, K.; Taur, S.; Jhaveri, T.; Dhali, S.; Chennareddy, S.; Potla, A.; Dikshit, J.B.; et al. Proteomic investigation of Falciparum and Vivax malaria for identification of surrogate protein markers. PLoS One 2012, 7, e41751. [Google Scholar]
- Ray, S.; Kamath, K.S.; Srivastava, R.; Raghu, D.; Gollapalli, K.; Jain, R.; Gupta, S.V.; Ray, S.; Taur, S.; Dhali, S.; et al. Serum proteome analysis of vivax malaria: An insight into the disease pathogenesis and host immune response. J. Proteomics 2012, 75, 3063–3080. [Google Scholar] [CrossRef]
- Saad, A.A.; Mohamed, O.E.; Ali, A.A.A.; Bashir, A.M.; Ali, N.I.A.; Elbashir, M.I.; Adam, I. Acute-phase proteins in pregnant Sudanese women with severe Plasmodium falciparum malaria. Trans. R. Soc. Trop. Med. Hyg. 2012, 106, 570–572. [Google Scholar] [CrossRef]
- Ho, M.; White, N.J. Molecular mechanisms of cytoadherence in malaria. Am. Physiol. Soc. 1999, 276, 1231–1242. [Google Scholar]
- De Kossodo, S.; Grau, G.E. Role of cytokines and adhesion molecules in malaria immunopathology. Stem Cells 1993, 11, 41–48. [Google Scholar] [CrossRef]
- Armah, H.; Dodoo, A.K.; Wiredu, E.K.; Stiles, J.K.; Adjei, A.A.; Gyasi, R.K.; Tettey, Y. High-level cerebellar expression of cytokines and adhesion molecules in fatal, paediatric, cerebral malaria. Ann. Trop. Med. Parasitol. 2005, 99, 629–647. [Google Scholar] [CrossRef]
- Sinha, S.; Qidwai, T.; Kanchan, K.; Anand, P.; Pati, G.N.S.J.S.; Mohanty, S.; Mishra, S.K.; Tyagi, P.K.; Sharma, S.K.; Consortium, I.G.V.; et al. Variations in host genes encoding adhesion molecules and susceptibility to falciparum malaria in India. Malar. J. 2008, 7. [Google Scholar] [CrossRef]
- Berendt, A.R.; Simmons, D.L.; Tansey, J.; Newbold, C.I.; Marsh, K. Intercellular adhesion molecule-1 is an endothelial cell adhesion receptor for Plasmodium falciparum. Nature 1989, 341, 57–59. [Google Scholar] [CrossRef]
- Clos, T.W.D. Function of C-reactive protein. Ann. Med. 2000, 4, 274–278. [Google Scholar] [CrossRef]
- Koenig, W.; Sund, M.; Fröhlich, M.; Fischer, H.; Löwe, H.; Döring, A.; Winston, L.; Hutchinson, M.B.P. C-Reactive Protein, a sensitive Marker of inflammation, Predicts Future Risk of Coronary Heart Disease in Initialyl Healthy Middle-Aged Men. Circulation 1999, 99, 237–242. [Google Scholar] [CrossRef]
- Hurt, N.; Smith, T.; Tanner, M.; Mwankusye, S.; Bordmann, G.; Weiss, N.A.; Teuscher, T. Evaluation of C-reactive protein and haptoglobin as malaria episode markers in an area of high transmission in Africa. Trans. R. Soc. Trop. Med. Hyg. 1994, 88, 182–186. [Google Scholar] [CrossRef]
- Nussler, A.; Pied, S.; Pontet, M.; Miltgen, F.; Renia, L.; Gentilini, M.; Mazier, D. Inflammatory status and preerythrocytic stages of malaria: Role of the C-reactive protein. Exp. Parasitol. 1991, 72, 1–7. [Google Scholar] [CrossRef]
- Dobryszycka, W. Biological functions of Haptoglobin-New pieces to an old puzzle. Clin. Chem. Lab. Med. 2009, 35, 647–654. [Google Scholar]
- Rogerson, S. What is the relationship between Haptoglobin, Malaria, and Anaemia? PLoS Med. 2006, 3, e200. [Google Scholar] [CrossRef]
- Imrie, H.; Fowkes, F.J.I.; Michon, P.; Tavul, L.; Hume, J.C.C.; Piper, K.P.; Reeder, J.C.; Day, K.P. Haptoglobin levels are associated with haptoglobin genotype and α+-Thalassemia in a malaria-endemic area. Am. J. Trop. Med. Hyg. 2006, 74, 965–971. [Google Scholar]
- Imrie, H.; Carter, M.; Hadjuk, S.; Day, K.P. Killing of Plasmodium falciparum by human serum haptoglobin. Mol. Biochem. Parasitol. 2004, 133, 93–98. [Google Scholar] [CrossRef]
- Singh, B.; Cox-Singh, J.; Miller, A.O.; Abdullah, M.S.; Snounou, G.; Rahman, H.A. Detection of malaria in Malaysia by nested polymerase chain reaction amplification of dried blood spots on filter papers. Trans. R. Soc. Trop. Med. Hyg. 1996, 90, 519–521. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mu, A.K.-W.; Bee, P.C.; Lau, Y.L.; Chen, Y. Identification of Protein Markers in Patients Infected with Plasmodium knowlesi, Plasmodium falciparum and Plasmodium vivax. Int. J. Mol. Sci. 2014, 15, 19952-19961. https://doi.org/10.3390/ijms151119952
Mu AK-W, Bee PC, Lau YL, Chen Y. Identification of Protein Markers in Patients Infected with Plasmodium knowlesi, Plasmodium falciparum and Plasmodium vivax. International Journal of Molecular Sciences. 2014; 15(11):19952-19961. https://doi.org/10.3390/ijms151119952
Chicago/Turabian StyleMu, Alan Kang-Wai, Ping Chong Bee, Yee Ling Lau, and Yeng Chen. 2014. "Identification of Protein Markers in Patients Infected with Plasmodium knowlesi, Plasmodium falciparum and Plasmodium vivax" International Journal of Molecular Sciences 15, no. 11: 19952-19961. https://doi.org/10.3390/ijms151119952




