Identification of Novel MicroRNAs in Primates by Using the Synteny Information and Small RNA Deep Sequencing Data
Abstract
:1. Introduction
2. Results and Discussion
2.1. Computational Identification of Marmoset Pre-miRNAs
2.2. Computational Identification of Pre-miRNAs in the Other Four Primates
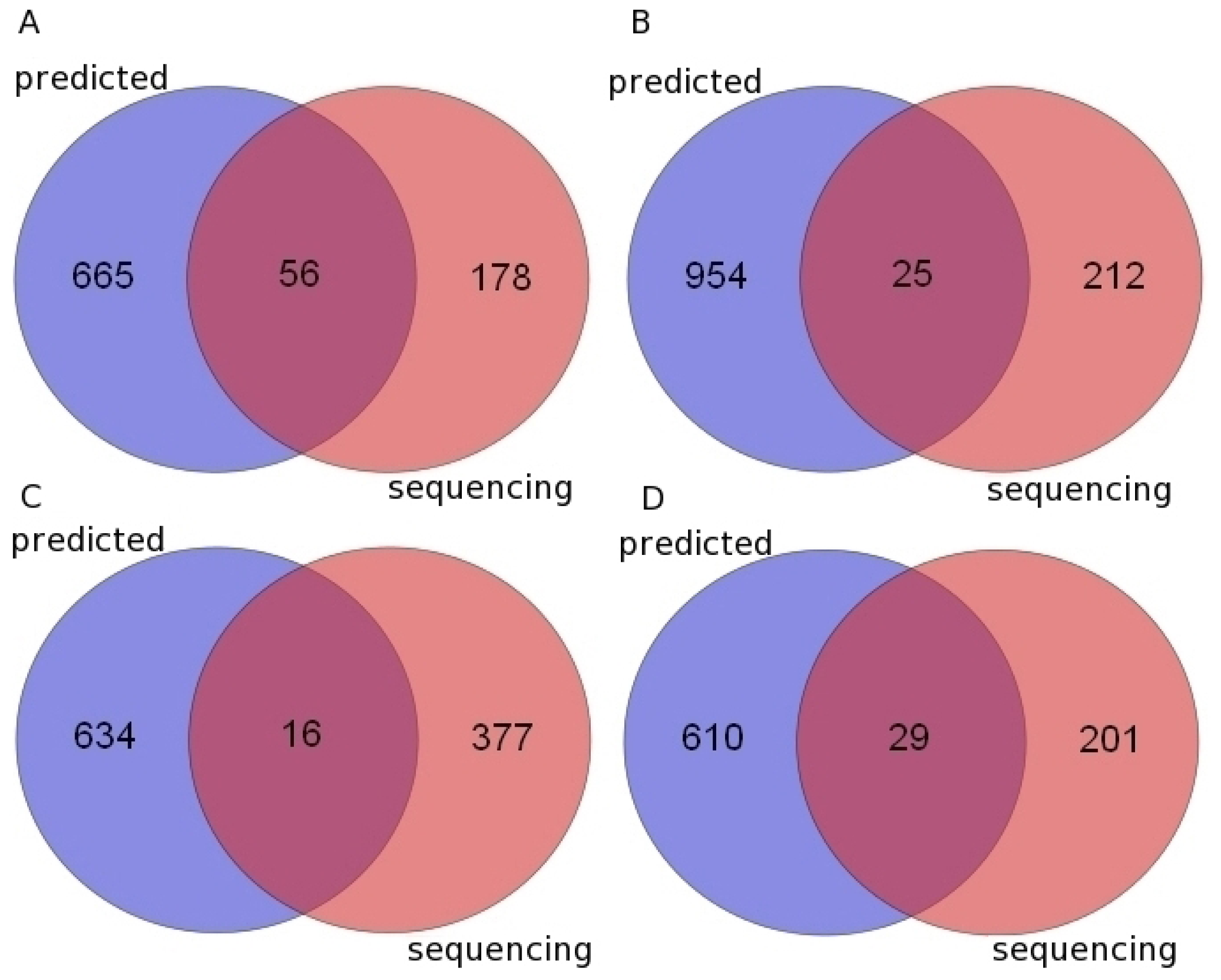
2.3. Novel Predicted Pre-miRNAs Validated by High Throughput Small RNA Sequencing Data
2.4. The Interesting Functional and Biological Importance of These Validated miRNAs
2.5. The Potential Transcription Factor Binding Sites of the Validated miRNA Genes Which Involve in Nervous System
3. Experimental Section
3.1. Data sources
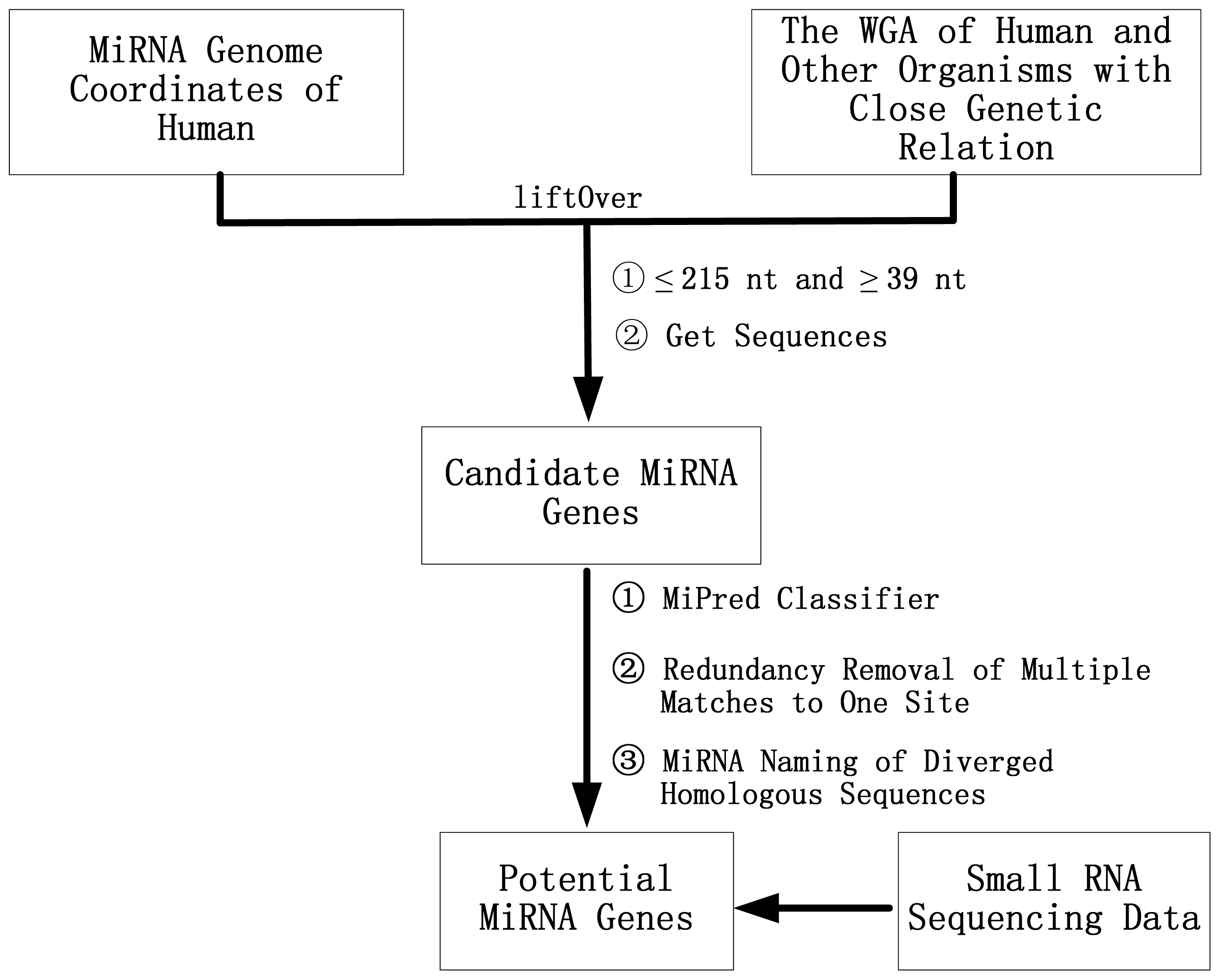
3.2. A Comparative Genomics Method for the Identification of Orthologous Pre-miRNAs
3.3. Deep Sequencing Data Analysis and New miRNAs Identification
3.4. Target Gene Ontology Analysis and Transcription Factors and Transcription Factor Binding Sites Exploration
4. Conclusions
| Primates | miRBase 19 (Known miRNA Genes) | Candidate miRNA Genes | Supported by Small RNA Sequencing Data | New miRNA Genes Identified from Deep Sequencing Data | |
|---|---|---|---|---|---|
| Re-identified miRNA Genes | Novel miRNA Genes | ||||
| Human | 1600 | ND | ND | ND | ND |
| Chimpanzee | 655 | 601 | 721 | 56 | 234 |
| Gorilla | 322 | 301 | 979 | 25 | 237 |
| Orangutan | 633 | 590 | 650 | 16 | 393 |
| Rhesus macaque | 535 | 480 | 639 | 29 | 230 |
| Marmoset | 0 | 0 | 929 | UA | UA |
| Chromosome | Start | End | Gene Symbol | Intron | Strand | Chromosome | Start | End | pre-miRNA | Strand |
|---|---|---|---|---|---|---|---|---|---|---|
| chr1 | 196871470 | 196873147 | PHC2 | intron_11 | + | chr1 | 196873044 | 196873102 | ppy-mir-3605 | + |
| chr7 | 130301326 | 130318076 | EXOC4 | intron_3 | + | chr7 | 130303121 | 130303184 | ppy-mir-4419a | + |
| chr19 | 51404019 | 51404127 | PTOV1 | intron_2 | + | chr19 | 51404069 | 51404128 | ppy-mir-4706 | + |
| chr20 | 13523168 | 13526970 | POLR3F | intron_2 | + | chr20 | 13524745 | 13524802 | ppy-mir-3192 | + |
| chr17_random | 2435896 | 2459049 | SKA2 | intron_1 | + | chr17_random | 2439995 | 2440051 | ppy-mir-301a | + |
| chr3 | 49052547 | 49062118 | SPINK8 | intron_2 | − | chr3 | 49059002 | 49059061 | ptr-mir-2115 | − |
| chr4 | 176472868 | 176516173 | GALNT7 | intron_2 | + | chr4 | 176492588 | 176492646 | ptr-mir-548t | + |
| chr11 | 65715927 | 65719260 | NDUFS8 | intron_6 | + | chr11 | 65716414 | 65716476 | ptr-mir-4691 | + |
| chr13 | 102152614 | 102687690 | FGF14 | intron_4 | − | chr13 | 102247593 | 102247653 | ptr-mir-2681 | − |
| chr15 | 63169799 | 63174675 | DENND4A | intron_21 | − | chr15 | 63171059 | 63171120 | ptr-mir-4511 | − |
| chr21 | 26267629 | 26354246 | DSCAM | intron_22 | − | chr21 | 26290766 | 26290830 | ptr-mir-4760 | − |
| chr22 | 44543820 | 44616196 | ATXN10 | intron_9 | + | chr22 | 44567304 | 44567367 | ptr-mir-4762 | + |
Acknowledgments
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar]
- Berezikov, E.; Cuppen, E.; Plasterk, R.H. Approaches to microRNA discovery. Nat. Genet 2006, 38, S2–S7. [Google Scholar]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol 2006, 57, 19–53. [Google Scholar]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol 2000, 7, 203–214. [Google Scholar]
- Artzi, S.; Kiezun, A.; Shomron, N. miRNAminer: A tool for homologous microRNA gene search. BMC Bioinforma 2008. [Google Scholar] [CrossRef]
- Zhou, M.; Wang, Q.; Sun, J.; Li, X.; Xu, L.; Yang, H.; Shi, H.; Ning, S.; Chen, L.; Li, Y.; et al. In silico detection and characteristics of novel microRNA genes in the Equus caballus genome using an integrated ab initio and comparative genomic approach. Genomics 2009, 94, 125–131. [Google Scholar]
- Brameier, M. Genome-wide comparative analysis of microRNAs in three non-human primates. BMC Res. Notes 2010. [Google Scholar] [CrossRef]
- Kent, W.J. BLAT—The BLAST-like alignment tool. Genome Res 2002, 12, 656–664. [Google Scholar]
- Baev, V.; Daskalova, E.; Minkov, I. Computational identification of novel microRNA homologs in the chimpanzee genome. Comput. Biol. Chem 2009, 33, 62–70. [Google Scholar]
- Weber, M.J. New human and mouse microRNA genes found by homology search. FEBS J 2005, 272, 59–73. [Google Scholar]
- Luo, Y.; Zhang, S. Computational prediction of amphioxus microRNA genes and their targets. Gene 2009, 428, 41–46. [Google Scholar]
- Meyer, L.R.; Zweig, A.S.; Hinrichs, A.S.; Karolchik, D.; Kuhn, R.M.; Wong, M.; Sloan, C.A.; Rosenbloom, K.R.; Roe, G.; Rhead, B.; et al. The UCSC Genome Browser database: Extensions and updates 2013. Nucleic Acids Res 2013, 41, D64–D69. [Google Scholar]
- Miller, W.; Rosenbloom, K.; Hardison, R.C.; Hou, M.; Taylor, J.; Raney, B.; Burhans, R.; King, D.C.; Baertsch, R.; Blankenberg, D.; et al. 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res 2007, 17, 1797–1808. [Google Scholar]
- Schwartz, S.; Kent, W.J.; Smit, A.; Zhang, Z.; Baertsch, R.; Hardison, R.C.; Haussler, D.; Miller, W. Human-mouse alignments with BLASTZ. Genome Res 2003, 13, 103–107. [Google Scholar]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.L.; Zhao, Y.; McDonald, H.; Zeng, T.; Hirst, M.; et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 2008, 18, 610–621. [Google Scholar]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar]
- Friedländer, M.R.; Adamidi, C.; Han, T.; Lebedeva, S.; Isenbarger, T.A.; Hirst, M.; Marra, M.; Nusbaum, C.; Lee, W.L.; Jenkin, J.C.; et al. High-resolution profiling and discovery of planarian small RNAs. Proc. Natl. Acad. Sci. USA 2009, 106, 11546–11551. [Google Scholar]
- Zhang, G.; Guo, G.; Hu, X.; Zhang, Y.; Li, Q.; Li, R.; Zhuang, R.; Lu, Z.; He, Z.; Fang, X.; et al. Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res 2010, 20, 646–654. [Google Scholar]
- Joyce, C.E.; Zhou, X.; Xia, J.; Ryan, C.; Thrash, B.; Menter, A.; Zhang, W.; Bowcock, A.M. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum. Mol. Genet 2012, 20, 4025–4040. [Google Scholar]
- Dhahbi, J.M.; Atamna, H.; Boffelli, D.; Magis, W.; Spindler, S.R.; Martin, D.I. Deep sequencing reveals novel microRNAs and regulation of microRNA expression during cell senescence. PLoS One 2011, 6, e20509. [Google Scholar]
- Sunkar, R.; Zhou, X.; Zheng, Y.; Zhang, W.; Zhu, J.K. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol 2008. [Google Scholar] [CrossRef]
- Jiang, P.; Wu, H.; Wang, W.; Ma, W.; Sun, X.; Lu, Z. MiPred: Classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res 2007, 35, W339–W344. [Google Scholar]
- Hofacker, I.L.; Fontana, W.; Stadler, P.F.; Bonhoeffer, L.S.; Tacker, M.; Schuster, P. Fast folding and comparison of RNA secondary structures. Chem. Mon 1994, 125, 167–188. [Google Scholar]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res 2006, 34, D140–D144. [Google Scholar]
- Piriyapongsa, J.; Jordan, I.K. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS One 2007, 2, e203. [Google Scholar]
- Yuan, Z.; Sun, X.; Liu, H.; Xie, J. MicroRNA genes derived from repetitive elements and expanded by segmental duplication events in mammalian genomes. PLoS One 2011, 6, e17666. [Google Scholar]
- Nawrocki, E.P.; Kolbe, D.L.; Eddy, S.R. Infernal 1.0: Inference of RNA alignments. Bioinformatics 2009, 25, 1335–1337. [Google Scholar]
- Dannemann, M.; Nickel, B.; Lizano, E.; Burbano, H.A.; Kelso, J. Annotation of primate miRNAs by high throughput sequencing of small RNA libraries. BMC Genomics 2012, 13. [Google Scholar] [CrossRef]
- Hu, H.Y.; Guo, S.; Xi, J.; Yan, Z.; Fu, N.; Zhang, X.; Menzel, C.; Liang, H.; Yang, H.; Zhao, M.; et al. MicroRNA expression and regulation in human, chimpanzee, and macaque brains. PLoS Genet 2011, 7, e1002327. [Google Scholar]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 2012, 40, 37–52. [Google Scholar]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar]
- Kertesz, M.; Iovino, N.; Unnerstall, U.; Gaul, U.; Segal, E. The role of site accessibility in microRNA target recognition. Nat. Genet 2007, 39, 1278–1284. [Google Scholar]
- Ozsolak, F.; Poling, L.L.; Wang, Z.; Liu, H.; Liu, X.S.; Roeder, R.G.; Zhang, X.; Song, J.S.; Fisher, D.E. Chromatin structure analyses identify miRNA promoters. Genes Dev 2008, 22, 3172–3183. [Google Scholar]
- Daenen, F.; van Roy, F.; de Bleser, P.J. Low nucleosome occupancy is encoded around functional human transcription factor binding sites. BMC Genomics 2008, 9. [Google Scholar] [CrossRef]
- Wang, J.; Zhuang, J.; Iyer, S.; Lin, X.; Whitfield, T.W.; Greven, M.C.; Pierce, B.G.; Dong, X.; Kundaje, A.; Cheng, Y.; et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res 2012, 22, 1798–1812. [Google Scholar]
- Kaplan, N.; Moore, I.K.; Fondufe-Mittendorf, Y.; Gossett, A.J.; Tillo, D.; Field, Y.; LeProust, E.M.; Hughes, T.R.; Lieb, J.D.; Widom, J.; et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 2009, 458, 362–366. [Google Scholar]
- Segal, E.; Fondufe-Mittendorf, Y.; Chen, L.; Thastrom, A.; Field, Y.; Moore, I.K.; Wang, J.P.; Widom, J. A genomic code for nucleosome positioning. Nature 2006, 442, 772–778. [Google Scholar]
- Portales-Casamar, E.; Thongjuea, S.; Kwon, A.T.; Arenillas, D.; Zhao, X.; Valen, E.; Yusuf, D.; Lenhard, B.; Wasserman, W.W.; Sandelin, A. JASPAR 2010: The greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res 2010, 38, D105–D110. [Google Scholar]
- Rhead, B.; Karolchik, D.; Kuhn, R.M.; Hinrichs, A.S.; Zweig, A.S.; Fujita, P.A.; Diekhans, M.; Smith, K.E.; Rosenbloom, K.R.; Raney, B.J.; et al. The UCSC Genome Browser database: Update 2010. Nucleic Acids Res 2010, 38, D613–D619. [Google Scholar]
- Gardner, P.P.; Daub, J.; Tate, J.; Moore, B.L.; Osuch, I.H.; Griffiths-Jones, S.; Finn, R.D.; Nawrocki, E.P.; Kolbe, D.L.; Eddy, S.R.; et al. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res 2011, 39, D141–D145. [Google Scholar]
- Falcon, S.; Gentleman, R. Using GOstats to test gene lists for GO term association. Bioinformatics 2007, 23, 257–258. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yuan, Z.; Liu, H.; Nie, Y.; Ding, S.; Yan, M.; Tan, S.; Jin, Y.; Sun, X. Identification of Novel MicroRNAs in Primates by Using the Synteny Information and Small RNA Deep Sequencing Data. Int. J. Mol. Sci. 2013, 14, 20820-20832. https://doi.org/10.3390/ijms141020820
Yuan Z, Liu H, Nie Y, Ding S, Yan M, Tan S, Jin Y, Sun X. Identification of Novel MicroRNAs in Primates by Using the Synteny Information and Small RNA Deep Sequencing Data. International Journal of Molecular Sciences. 2013; 14(10):20820-20832. https://doi.org/10.3390/ijms141020820
Chicago/Turabian StyleYuan, Zhidong, Hongde Liu, Yumin Nie, Suping Ding, Mingli Yan, Shuhua Tan, Yuanchang Jin, and Xiao Sun. 2013. "Identification of Novel MicroRNAs in Primates by Using the Synteny Information and Small RNA Deep Sequencing Data" International Journal of Molecular Sciences 14, no. 10: 20820-20832. https://doi.org/10.3390/ijms141020820





