Molecular Cloning, Sequence Characterization and Expression Analysis of a CD63 Homologue from the Coleopteran Beetle, Tenebrio molitor
Abstract
:1. Introduction
2. Results
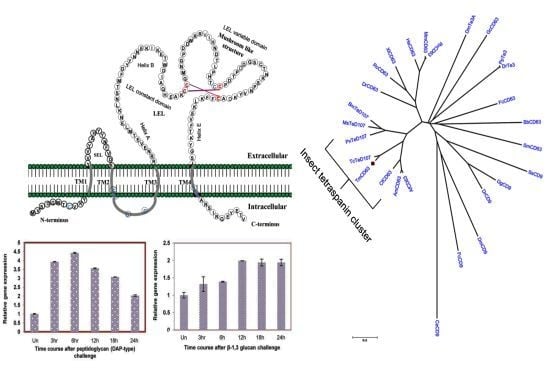
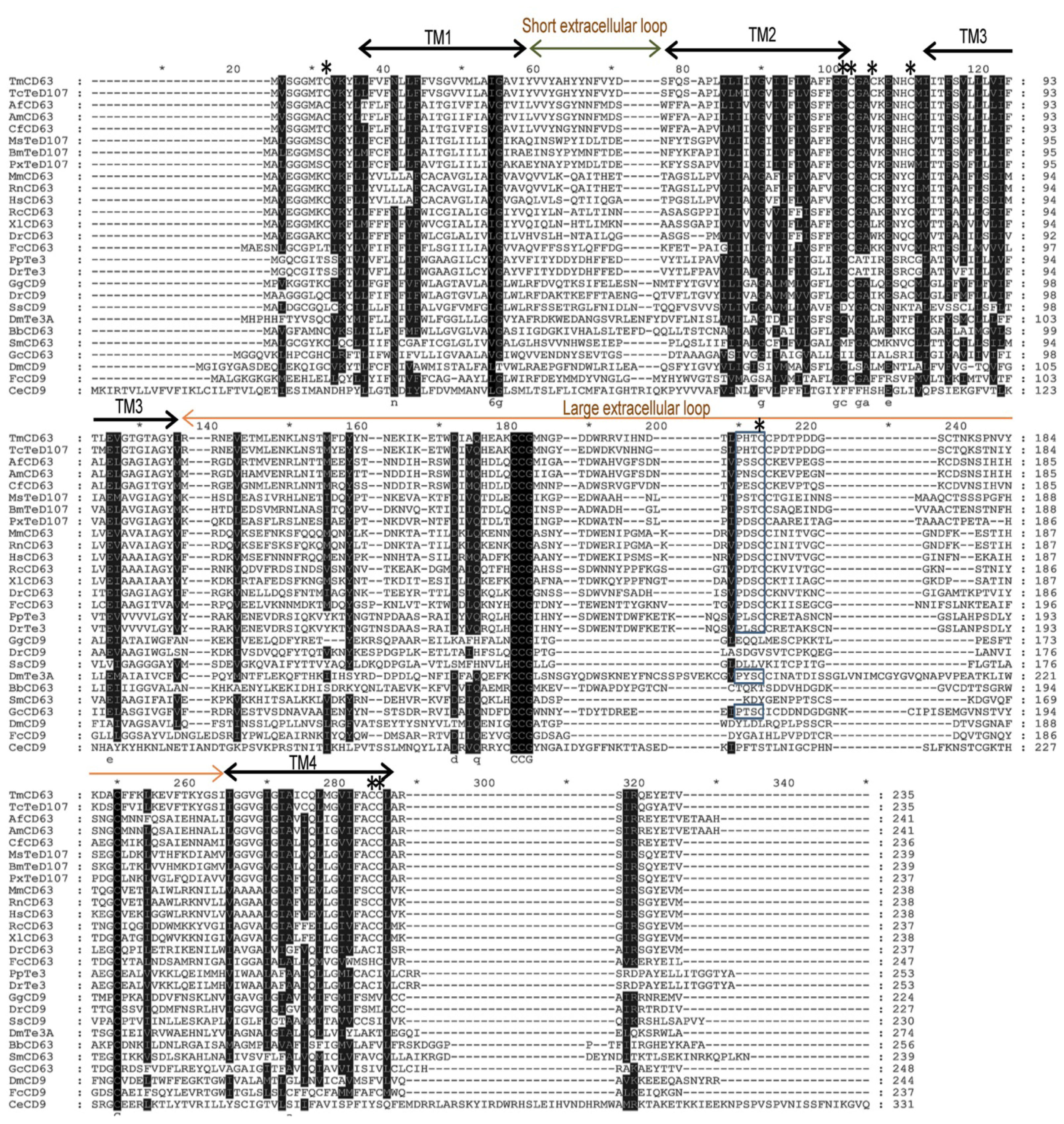
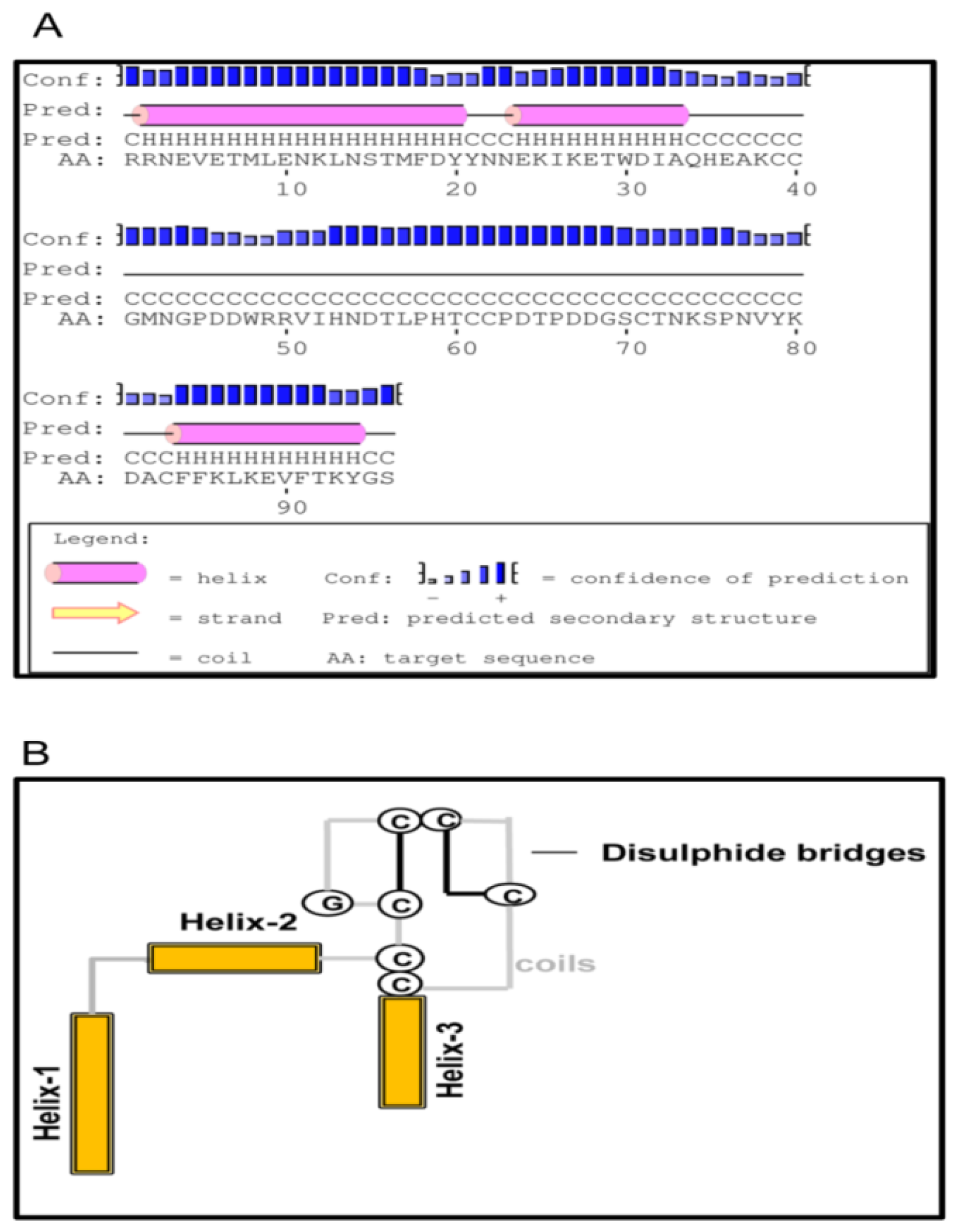
2.1. Bioinformatics Analysis of TmCD63 and Structure of Deduced Protein
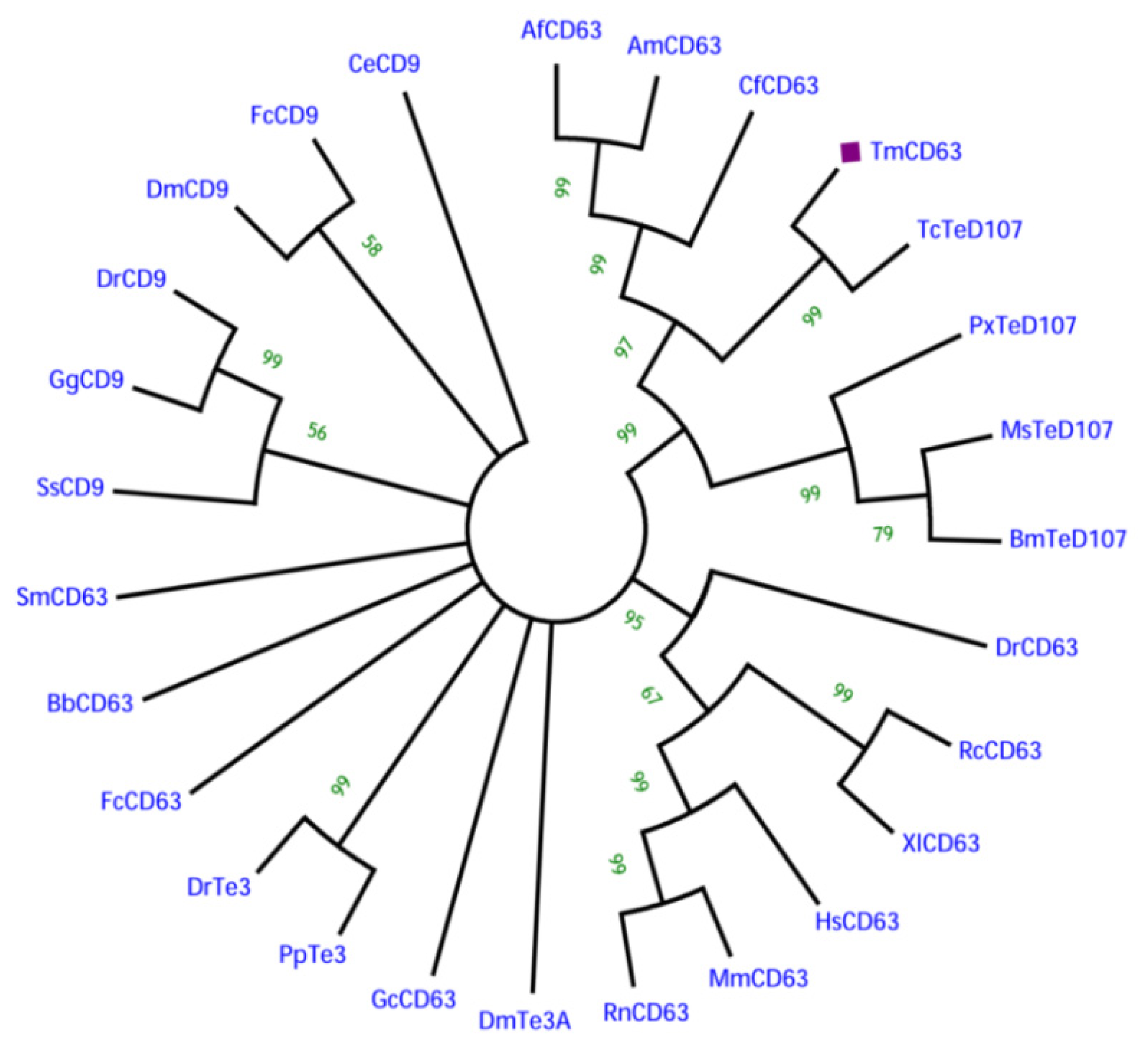
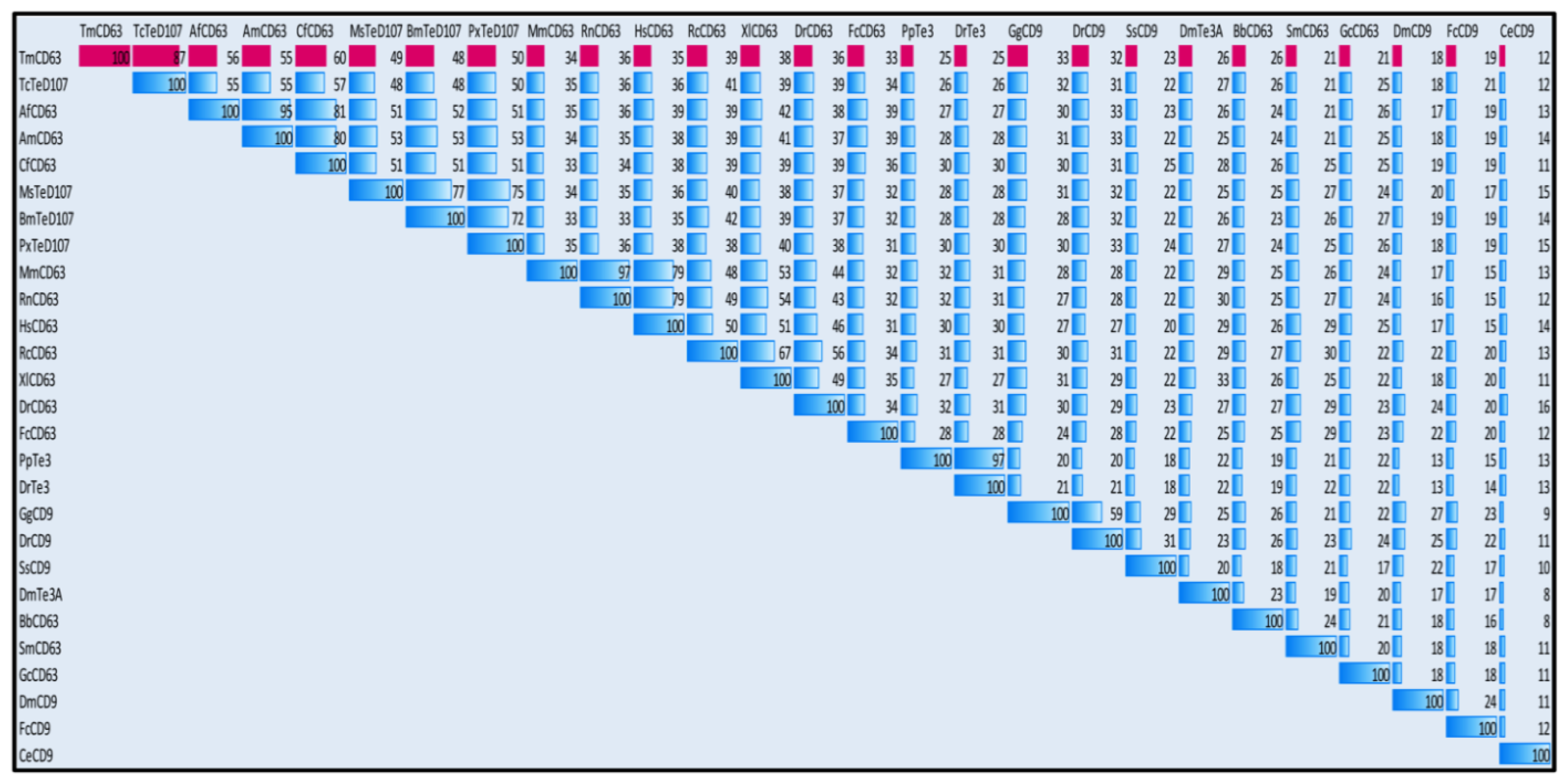
2.2. Phylogenetic Analysis of TmCD63 and Secondary Structure Prediction
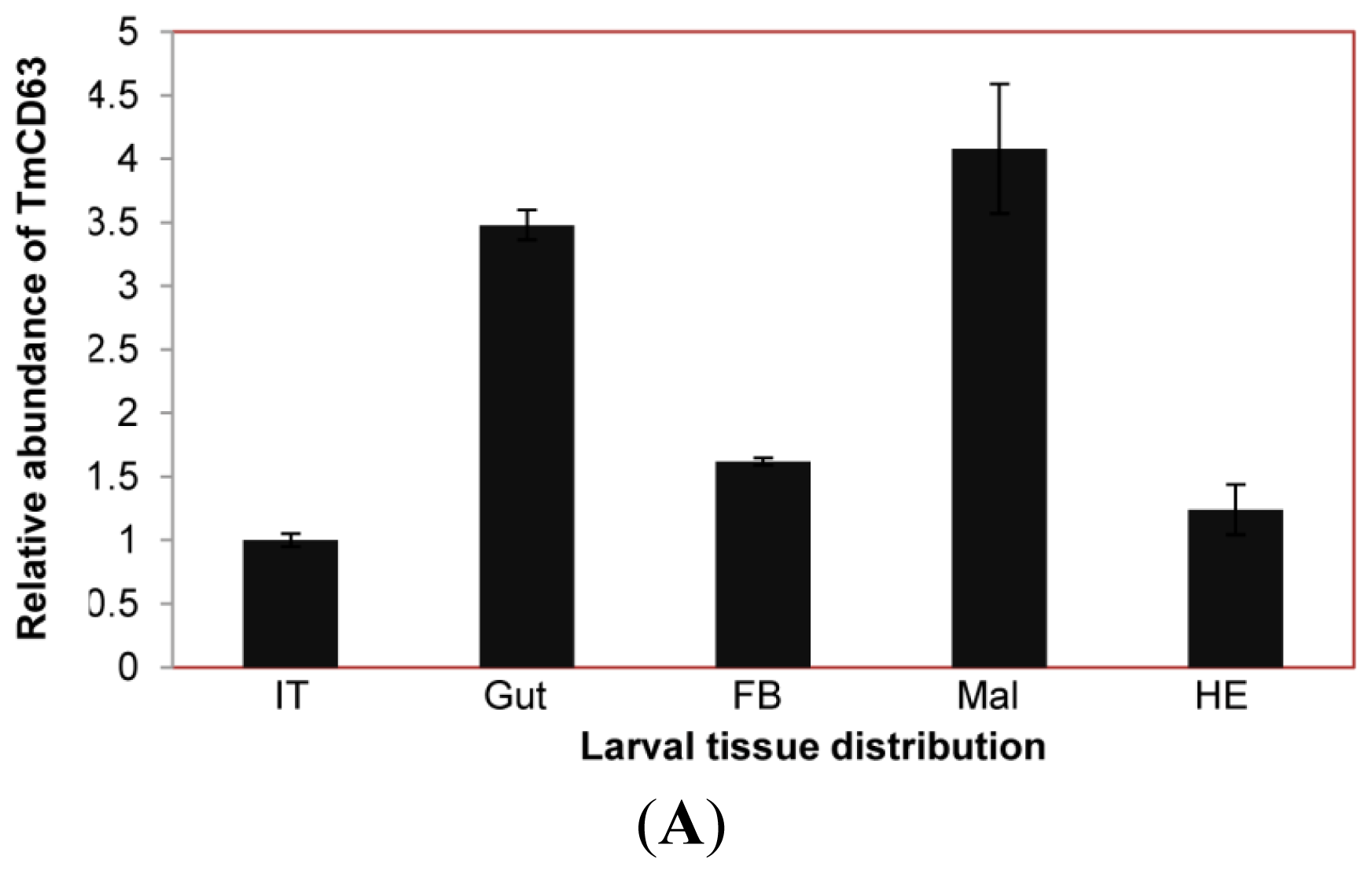
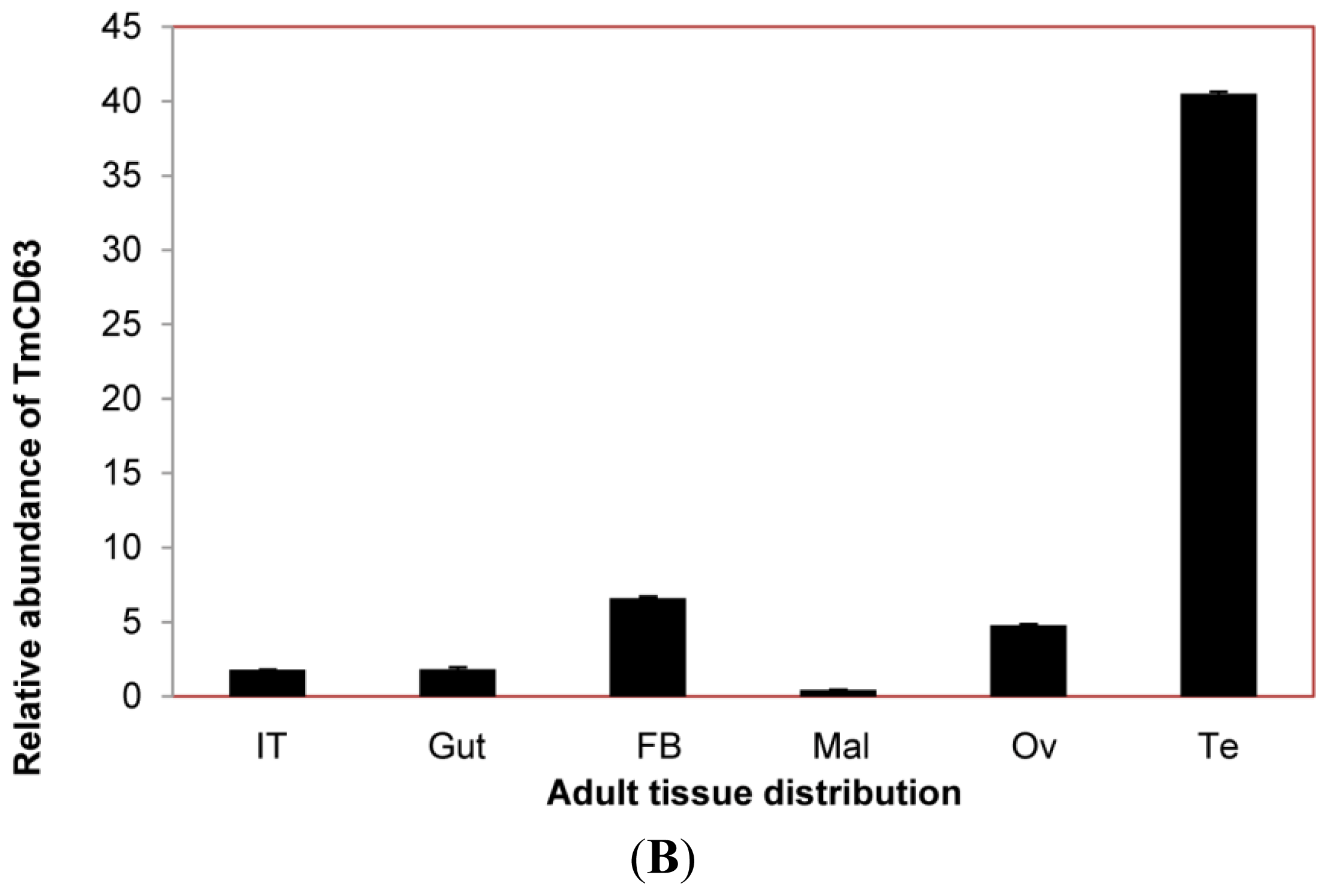
2.3. Developmental and Tissue Distribution of TmCD63
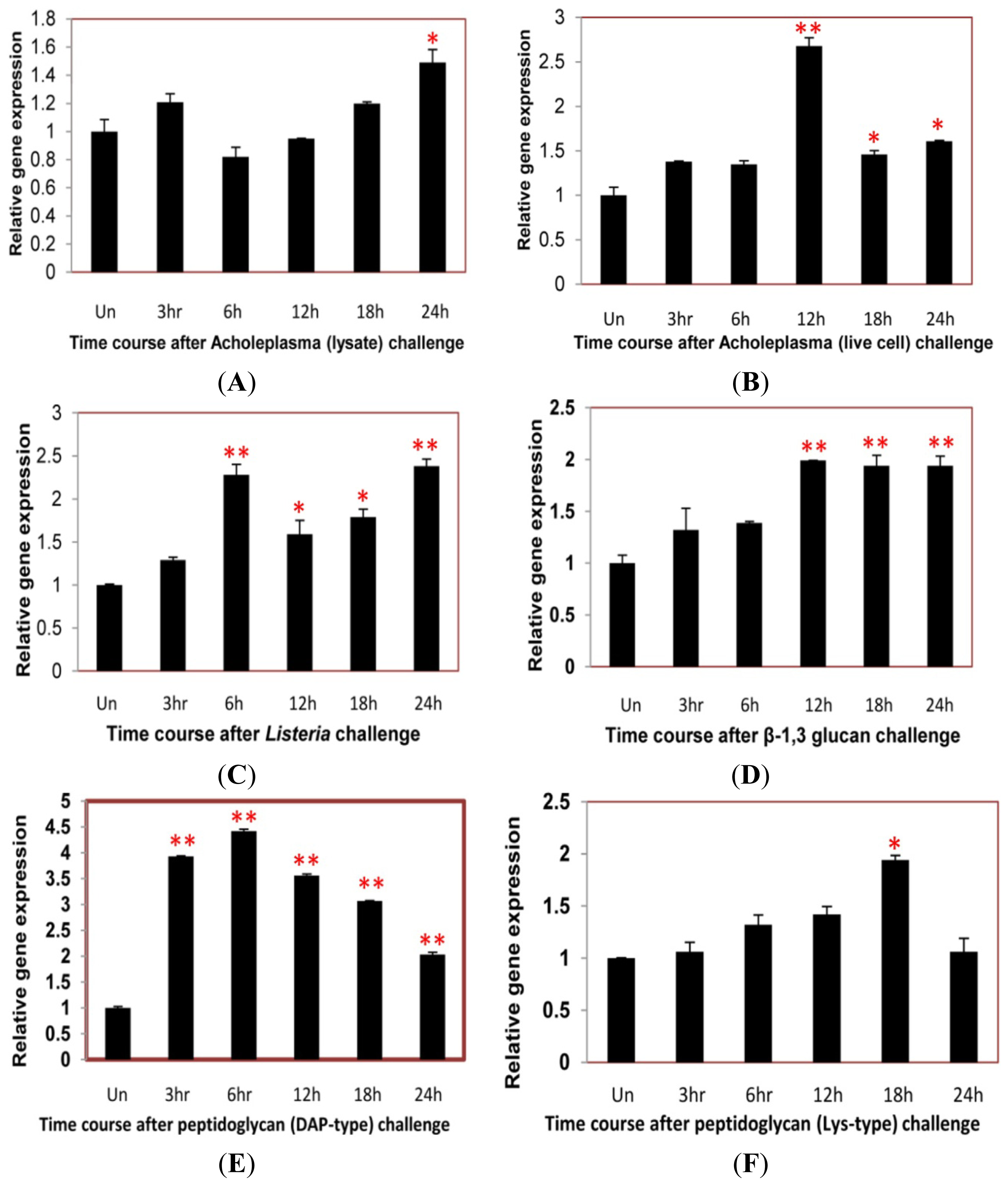
2.4. Time Course Analysis of TmCD63 after Immune Elicitor Challenge
3. Discussion
4. Experimental Section
4.1. Insect Rearing
4.2. Chemicals and Strains
4.3. Construction of cDNA Library of T. molitor Larvae
4.4. Cloning of the Full-Length cDNA of TmCD63
4.5. TmCD63 Sequence and Phylogenetic Analysis
4.6. Tissue Distribution and Developmental Expression Profile of TmCD63 by Quantitative Real-Time PCR
4.7. Immune Elicitors and Challenge Studies
4.8. Time-Course Analysis of TmCD63 by Quantitative Real-Time PCR
5. Conclusions
Supplementary Information
ijms-14-20744-s001.pdf
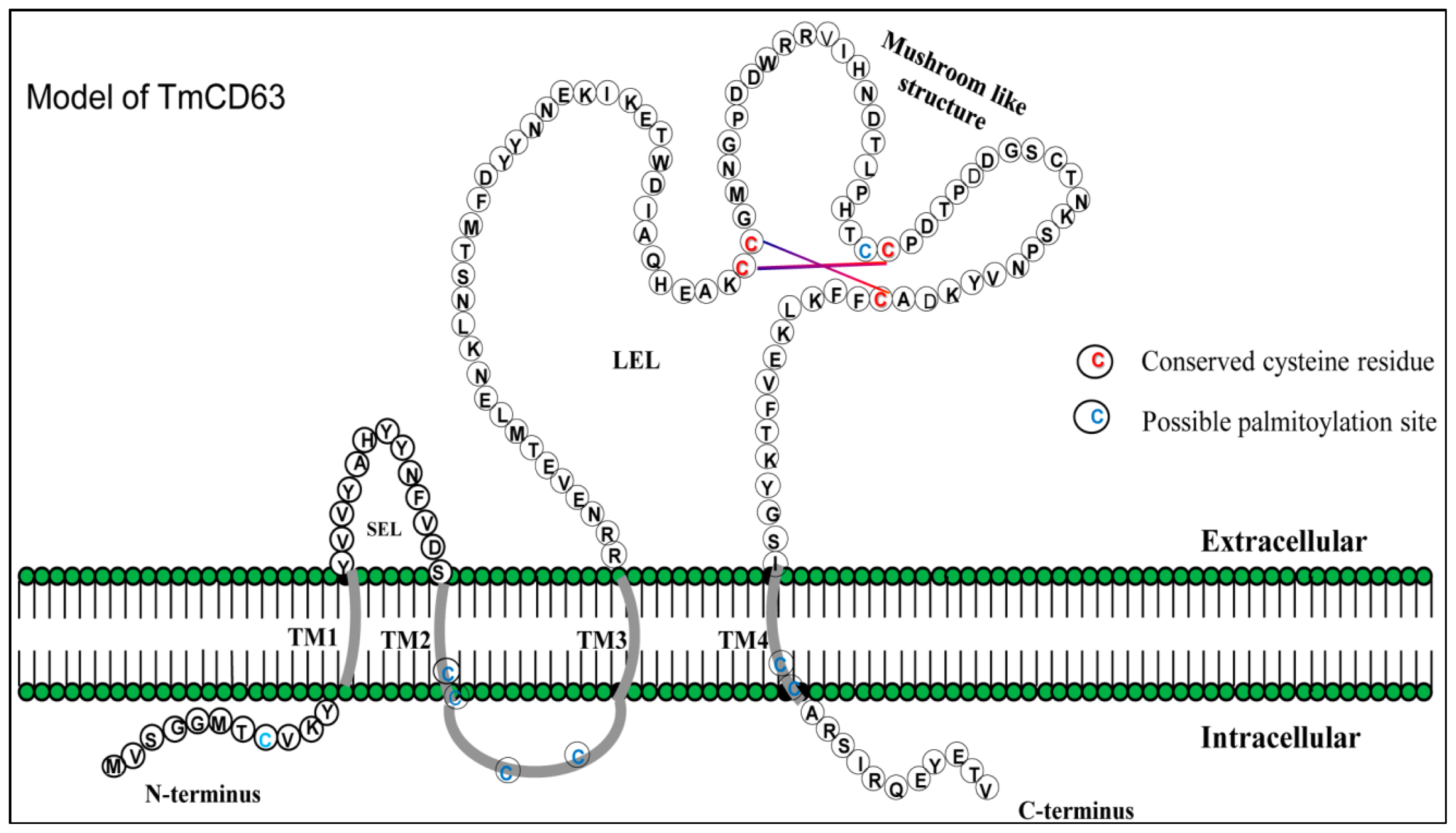

| Position | Peptide | Score | Cutoff | Cluster |
|---|---|---|---|---|
| 8 | MVSGGMTCVKYLLFV | 0.595 | 0.308 | Cluster A |
| 71 | FLVAFFGCCGACKEN | 1.694 | 0.497 | Cluster B |
| 72 | LVAFFGCCGACKENH | 2.278 | 0.497 | Cluster B |
| 75 | FFGCCGACKENHCMI | 0.852 | 0.497 | Cluster B |
| 80 | GACKENHCMIITFSV | 2.322 | 1.225 | Cluster C |
| 166 | NDTLPHTCCPDTPDD | 0.806 | 0.497 | Cluster B |
| 222 | LMGVIFACCLARSIR | 3.433 | 0.308 | Cluster A |
| 223 | MGVIFACCLARSIRQ | 3.481 | 0.497 | Cluster B |
Acknowledgments
Conflicts of Interest
References
- Oren, R.; Takahashi, S.; Doss, C.; Levy, R.; Levy, S. TAPA-1, the target of an antiproliferative antibody, defines a new family of transmembrane proteins. Mol. Cell Biol 1990, 10, 4007–4015. [Google Scholar]
- Metzelaar, M.J.; Wijngaard, P.L.; Peters, P.J.; Sixma, J.J.; Nieuwenhuis, H.K.; Clevers, H.C. CD63 antigen. A novel lysosomal membrane glycoprotein, cloned by a screening procedure for intracellular antigens in eukaryotic cells. J. Biol. Chem 1991, 266, 3239–3245. [Google Scholar]
- Maecker, H.T.; Todd, S.C.; Levy, S. The tetraspanin superfamily: Molecular facilitators. FASEB J 1997, 11, 428–442. [Google Scholar]
- Todres, E.; Nardi, J.B.; Robertson, H.M. The tetraspanin superfamily in insects. Insect Mol. Biol 2000, 9, 581–590. [Google Scholar]
- Kovalenko, O.V.; Yang, X.; Kolesnikova, T.V.; Hemler, M.E. A novel cysteine crosslinking method reveals a direct association between claudin-1 and tetraspanin CD9. Mol. Cell Prot 2007, 6, 1855–1867. [Google Scholar]
- DeSalle, R.; Mares, R.; Garcia-Espana, A. Evolution of cysteine patterns in the large extracellular loop of tetraspanins from animals, fungi, plants and single-celled eukaryotes. Mol. Phylogenet. Evol 2010, 56, 486–491. [Google Scholar]
- Hemler, M.E. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell. Dev. Biol 2003, 19, 397–422. [Google Scholar]
- Martin, F.; Roth, D.M.; Jans, D.A.; Pouton, C.W.; Partridge, L.J.; Monk, P.N.; Moseley, G.W. Tetraspanins in viral infections: A Fundamental role in viral biology? J. Virol 2005, 79, 10839–10851. [Google Scholar]
- Levy, S.; Shoham, T. The tetraspanin web modulates immune-signaling complexes. Nat. Rev. Immunol 2005, 5, 136–148. [Google Scholar]
- Nishiuchi, R.; Sanzen, N.; Nada, S.; Sumida, Y.; Wada, Y.; Okada, M.; Takagi, J.; Hasegawa, H.; Sekiguchi, K. Potentiation of the ligand-binding activity of integrin α3β1 via association with tetraspanin CD151. Proc. Natl. Acad. Sci. USA 2005, 102, 1939–1944. [Google Scholar]
- Berditchevski, F.; Odintsova, E. Tetraspanins as regulators of protein trafficking. Traffic 2007, 8, 89–96. [Google Scholar]
- Pols, M.S.; Klumperman, J. Trafficking and function of the tetraspanin CD63. Expt. Cell Res 2009, 315, 1584–1592. [Google Scholar]
- Kovalenko, O.V.; Yang, X.; Kolesnikova, T.V.; Hemler, M.E. Evidence for specific tetraspanin homodimers: Inhibition of palmitoylation makes cysteine residues available for cross-linking. Biochem. J 2004, 377, 407–417. [Google Scholar]
- Raposo, G.; Marks, M.S.; Cutler, D.F. Lysosome-related organelles: Driving post-Golgi compartments into specialization. Curr. Opin. Cell Biol 2007, 19, 394–401. [Google Scholar]
- Skubitz, K.M.; Campbell, K.D.; Iida, J.; Skubitz, A.P.N. CD63 associates with tyrosine kinase activity and CD11/CD18, and transmits an activation signal in neutrophils. J. Immunol 1996, 157, 3617–3626. [Google Scholar]
- Yoshida, T.; Kawano, Y.; Sato, K.; Ando, Y.; Aoki, J. A CD63 mutant inhibits T-cell tropic human immunodeficiency virus type-1 entry by disrupting CXCR4 trafficking to the plasma membrane. Traffic 2008, 9, 540–558. [Google Scholar]
- Hirst, J.; Bright, N.A.; Rous, B.; Robinson, M.S. Characterization of a fourth adaptor-related protein complex. Mol. Biol. Cell 1999, 10, 2787–2802. [Google Scholar]
- Lekishvili, T.; Fromm, E.; Mujoomdar, M.; Berditchevski, F. The tumor-associated antigen L6 (L6-Ag) is recruited to the tetraspanin-enriched microdomains: Implication for tumor cell motility. J. Cell Sci 2008, 121, 685–694. [Google Scholar]
- Latysheva, N.; Muratov, G.; Rajesh, S.; Padgett, M.; Hotchin, N.A.; Overduin, M.; Berditchevski, F. Syntenin-1 is a new component of tetraspanin-enriched microdomains: Mechanisms and consequences of the interaction of syntenin-1 with CD63. Mol. Cell Biol 2006, 26, 7707–7718. [Google Scholar]
- Jung, K.K.; Liu, X.W.; Chirco, R.; Fridman, R.; Kim, H.R.C. Identification of CD63 as a tissue inhibitor of metalloproteinase-1 interacting cell surface protein. EMBO J 2006, 25, 3934–3942. [Google Scholar]
- Duffield, A.; Karmsteeg, E.J.; Brown, A.N.; Pagel, P.; Caplan, M.J. The tetraspanin CD63 enhances the internalization of H, K-ATPase β-subunit. Proc. Natl. Acad. Sci. USA 2003, 100, 15560–15565. [Google Scholar]
- Takino, T.; Miyamori, H.; Kawaguchi, N.; Uekita, T.; Seiki, M.; Sato, H. Tetraspanin CD63 promotes targeting and lysosomal proteolysis of membrane-type 1matrix metalloproteinase. Biochem. Biophys. Res. Commun 2003, 304, 160–166. [Google Scholar]
- Hemler, M.E.; Mannion, B.A.; Berditchevski, F. Association of TM4SF proteins with integrins: Relevance to cancer. Biochem. Biophys. Acta 1996, 1287, 67–71. [Google Scholar]
- Azorsa, D.O.; Hyman, J.A.; Hildreth, J.E. CD63/Pltgp40: A platelet activation antigen identical to the stage-specific, melanoma-associated antigen ME491. Blood 1991, 78, 280–284. [Google Scholar]
- Koyama, Y.; Suzuki, M.; Yoshida, T. CD63, a member of tetraspanin transmembrane protein family, induces cellular spreading by reaction with monoclonal antibody on substrata. Biochem. Biophys. Res. Commun 1998, 246, 841–846. [Google Scholar]
- Heijnen, H.F.; Debili, N.; Vainchencker, W.; Breton-Gorius, J.; Geuze, H.J.; Sixma, J.J. Multivesicular bodies are an intermediate stage in the formation of platelet alpha-granules. Blood 1998, 91, 2313–2325. [Google Scholar]
- Kobayashi, T.; Vischer, U.M.; Rosnoblet, C.; Lebrand, C.; Lindsay, M.; Parton, R.G.; Kruithof, K.O.; Gruenberg, J. The tetraspanin CD63/lamp3 cycles between endocytic and secretory compartments in human endothelial cells. Mol. Biol. Cell 2000, 11, 1829–1843. [Google Scholar]
- Van der Wal, J.E.; Hermsen, M.A.J.A.; Gille, H.J.P.; Schouten-van Meeteren, N.Y.N.; Moll, A.C.; Imhof, S.M.; Meijer, G.A.; Baak, J.P.A.; van der Valk, P. Comparative genomic hybridization divides retinoblastomas into a high and a low level chromosomal instability group. J. Clin. Pathol 2003, 56, 26–30. [Google Scholar]
- Schroder, J.; Lullmann-Rauch, R.; Himmerkus, N.; Pleines, I.; Nieswandt, B. Deficiency of the tetraspanin CD63 associated with kidney pathology but normal lysosomal function. Mol. Cell. Biol 2009, 29, 1083–1094. [Google Scholar]
- Lin, D.; Kamsteeg, E.J.; Zhang, Y.; Jin, Y.; Sterling, H.; Yue, P.; Roos, M.; Duffield, A.; Spencer, J.; Caplan, M.; et al. Expression of tetraspanin protein CD63 activates protein-tyrosine kinase (PTK) and enhances the PTK-induced inhibition of ROMK channels. J. Biol. Chem 2008, 283, 7674–7681. [Google Scholar]
- Li, G.; Dziuba, N.; Friedrich, B.; Murray, J.L.; Ferguson, M.R. A post-entry role for CD63 in early HIV-1 replication. Virology 2011, 412, 315–324. [Google Scholar]
- Verweij, F.J.; Eijndhoven, M.; Hopmans, E.S.; Vendrig, T.; Wurdinger, T.; Cahir-McFarland, E.; Kieff, E.; Geerts, D.; Kant, R.; Neefjes, J.; et al. LMP-1 association with CD63 in endosomes and secretion via exosomes limits constitutive NK-κB activation. EMBO J 2011, 30, 2115–2129. [Google Scholar]
- Sordat, I.; Decraene, C.; Silvestre, T.; Petermann, O.; Auffray, C.; Pietu, G.; Sordat, B. Complimentary DNA arrays identify CD63 tetraspanin and alpha3 integrin chain as differentially expressed in low and high metastatic human colon carcinoma cells. Lab. Invest 2002, 82, 1715–1724. [Google Scholar]
- Jang, H.I.; Lee, H. A decrease in the expression of CD63 tetraspanin protein elevates invasive potential of human melanoma cells. Exp. Mol. Med 2003, 35, 317–323. [Google Scholar]
- Yeh, H.Y.; Klesius, P.H. Channel catfish (Ictalurus. punctatus, 1818) tetraspanin membrane protein family: Identification, characterization and expression analysis of CD63 cDNA. Vet. Immunol. Immunopathol 2010, 133, 302–308. [Google Scholar]
- Liu, Z.; Zhang, S.; Li, H.; Luan, J.; Wang, Y.; Wang, L.; Xiang, J. Characterization and expression profile of AmphiCD63 encoding a novel member of TM4SF proteins from amphioxus Branchiostoma belcheri tsingtauense. DNA Seq 2005, 16, 195–201. [Google Scholar]
- Wang, B.; Li, F.; Xiang, J.; Gui, L.; Luo, Z.; Yan, H. Three tetraspanins from Chinese shrimp, Fenneropenaeus chinensis, may play important roles in WSSV infection. J. Fish. Dis 2010, 33, 15–29. [Google Scholar]
- Kim, C.H.; Kim, S.J.; Kan, H.; Kwon, H.M.; Roh, K.B.; Jiang, R.; Yang, Y.; Park, J.W.; Lee, H.H.; Ha, N.C.; et al. A three-step proteolytic cascade mediates the activation of the peptidoglycan-induced toll pathway in an insect. J. Biol. Chem 2008, 283, 7599–7607. [Google Scholar]
- Jiang, R.; Kim, E.H.; Gong, J.H.; Kwon, H.M.; Kim, C.H.; Ryu, K.H.; Park, J.W.; Kurokawa, K.; Zhang, J.; Gubb, D.; et al. Three pairs of protease-serpin complexes cooperatively regulate the insect immune responses. J. Biol. Chem 2009, 284, 35652–35658. [Google Scholar]
- Zhu, J.Y.; Yang, P.; Zhang, Z.; Wu, G.X.; Yang, B. Transcriptomic immune response of Tenebrio molitor pupae to parasitization by Scleroderma guani. PLoS One 2013, 8, e54411. [Google Scholar]
- Jeong, J.E.; Kang, S.W.; Hwang, H.J.; Chae, S.W.; Patnaik, B.B.; Han, Y.S.; Lee, J.B.; Jo, Y.H.; Lee, B.L.; Seog, D.H.; et al. Expressed sequence tags (ESTs) analysis of Tenebrio molitor larvae. Entom. Res 2013, 43, 162–170. [Google Scholar]
- Adell, T.; Gamulin, V.; Perovic-Ottstadt, S.; Wiens, M.; Korzhev, M.; Muller, I.M.; Muller, W.E.G. Evolution of metazoan cell junction proteins: The scaffold protein MAGI and the transmembrane receptor tetraspanin in the demosponge Suberites domuncula. J. Mol. Evol 2004, 59, 41–50. [Google Scholar]
- Seigneuret, M.; Delaguillaumie, A.; Lagaudriere-Gesbert, C.; Conjeaud, H. Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J. Biol. Chem 2001, 276, 40055–40064. [Google Scholar]
- Kitadokoro, K.; Bordo, D.; Galli, G.; Petracca, R.; Falugi, F.; Abrignani, S.; Grandi, G.; Bolognesi, M. CD81 extracellular domain 3D structure: Insight into the tetraspanin superfamily structural motifs. EMBO J 2001, 20, 12–18. [Google Scholar]
- Levy, S.; Todd, S.C.; Maecker, H.T. CD81 (TAPA-1): A molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol 1998, 16, 89–109. [Google Scholar]
- Hemler, M.E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol 2005, 6, 801–811. [Google Scholar]
- Yang, X.; Claas, C.; Kraeft, S.-K.; Chen, L.B.; Wang, Z.; Kreidberg, J.A; Hemler, M.A. Palmitoylation of tetraspanin proteins: Modulation of CD151 lateral interactions, subcellular distribution and integrin-dependent cell morphology. Mol. Biol. Cell 2002, 13, 767–781. [Google Scholar]
- Hemler, M.E. Integrin-associated proteins. Curr. Opin. Cell Biol 1998, 10, 578–585. [Google Scholar]
- Flannery, A.R.; Czibener, C.; Andrews, N.W. Palmitoylation-dependent association with CD63 targets the Ca2+ sensor synaptotagmins VII to lysosomes. J. Cell Biol 2010, 191, 599–613. [Google Scholar]
- Bonifacino, J.S.; Traub, L.M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem 2003, 72, 395–447. [Google Scholar]
- Tang, W.; Kim, T.W.; Oses-Prieto, J.A.; Sun, Y.; Deng, Z.; Zhu, S.; Wang, R.; Burlingame, A.L.; Wang, Z.Y. Brassinosteroid-signalling kinases (BSKs) mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 2008, 321, 557–560. [Google Scholar]
- Baldauf, S.L. The deep roots of eukaryotes. Science 2003, 300, 1703–1706. [Google Scholar]
- Lambou, K.; Tharreau, D.; Kohler, A.; Sirven, C.; Marguerettaz, M.; Barbisan, C.; Sexton, A.C.; Kellner, E.M.; Martin, F.; Howlett, B.J.; et al. Fungi have three tetraspanin families with distinct functions. BMC Genomics 2008, 9. [Google Scholar] [CrossRef]
- Garcia-Espana, A.; Mares, R.; Sun, T.T.; DeSalle, R. Intron evolution: Testing hypothesis of intron evolution using the phylogenomics of tetraspanins. PLoS One 2009, 4, e4680. [Google Scholar]
- Muller, W.E.G.; Wimmer, W.; Schatton, W.; Bohm, M.; Batel, R.; Filie, Z. Initiation of an aquaculture of sponges for the sustainable production of bioactive metabolites in open systems: Example Geodia cydonium. Mar. Biotechnol 1999, 1, 569–579. [Google Scholar]
- Tham, T.N.; Gouin, E.; Rubinstein, E.; Boucheix, C.; Cossart, P.; Pizzaro-Cerda, J. Tetraspanin CD81 is required for Listeria monocytogenes invasion. Infect. Immun 2010, 78, 204–209. [Google Scholar]
- Pileri, P.; Uematsu, Y.; Campagnoli, S.; Galli, G.; Falugi, F.; Petracca, R.; Weiner, A.J.; Houghton, M.; Rosa, D.; Grandi, G.; et al. Binding of hepatitis C virus to CD81. Science 1998, 282, 938–941. [Google Scholar]
- Artavanis-Tsakonas, K.; Kasperkovitz, P.V.; Papa, E.; Cardenas, M.L.; Khan, N.S.; van der Veen, A.G.; Ploegh, H.L.; Vyas, J.M. The tetraspanin CD82 is specifically recruited to fungal and bacterial phagosomes prior to acidification. Infect. Immun 2011, 79, 1098–1106. [Google Scholar]
- Artavanis-Tsakonas, K.; Love, J.C.; Ploegh, H.L.; Vyas, J.M. Recruitment of CD63 to Cryptococcus neoformans phagosomes requires acidification. Proc. Natl. Acad. Sci. USA 2006, 103, 15945–15950. [Google Scholar]
- Green, L.R.; Monk, P.N.; Partridge, L.J.; Morris, P.; Gorringe, A.R.; Read, R.C. Cooperative role for tetraspanins in adhesion-mediated attachment of bacterial species to human epithelial cells. Infect. Immun 2011, 79, 2241–2249. [Google Scholar]
- Moribe, H.; Yochem, J.; Yamada, H.; Tabuse, Y.; Fujimoto, T.; Mekada, E. Tetraspanin protein (TSP-15) is required for epidermal integrity in Caenorhabditis elegans. J. Cell Sci 2004, 117, 5209–5220. [Google Scholar]
- McGonigle, L.; Mousley, A.; Marks, N.J.; Brennan, G.P.; Dalton, J.P.; Spithill, T.W.; Day, T.A.; Maule, A.G. The silencing of cysteine proteases in Fasciola hepatica newly excysted juveniles using RNA interference reduces gut penetration. Int. J. Parasitol 2008, 38, 149–155. [Google Scholar]
- National Center for Biotechnology Information. Available online: www.ncbi.nlm.nih.gov/gorf/gorf.html/ (accessed on 4 May 2013).
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acids Res 1997, 25, 4876–4882. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol. Biol. Evol 2011, 30. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol 1987, 4, 406–425. [Google Scholar]
- Ren, J.; Wen, L.; Gao, X.; Jin, C.; Xue, Y.; Yao, X. CSS Palm 3.0: An updated software for palmitoylation sites prediction. Prot. Engineer. Des. Select 2008, 21, 639–644. [Google Scholar]
- Petersen, T.N.; Brunak, S.; Heijne, G.; Nielsen, H. Signal 4.0: Discriminating signal peptides from transmembrane regions. Nature Methods 2011, 8, 785–786. [Google Scholar]
- ProtScale Tool. Available online: web expasy.org/protscale/ (accessed on 19 May 2013).
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol 1982, 5, 105–132. [Google Scholar]
- ProtParam Tool. Available online: web.expasy.org/protparam/ (accessed on 19 May 2013).
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 21–34. [Google Scholar]
- Bachere, E.; Chagot, D.; Grizel, H. Separation of Crassostrea gigas hemocytes by density gradient centrifugation and counterflow centrifugal elutriation. Dev. Comp. Immunol 1988, 12, 549–559. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar]
- Lee, N.S.; Dohjima, T.; Bauer, G.; Li, H.; Li, M.J.; Ehsani, A.; Salvaterra, P.; Rossi, J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol 2002, 20, 500–505. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Patnaik, B.B.; Kang, S.M.; Seo, G.W.; Lee, H.J.; Patnaik, H.H.; Jo, Y.H.; Tindwa, H.; Lee, Y.S.; Lee, B.L.; Kim, N.J.; et al. Molecular Cloning, Sequence Characterization and Expression Analysis of a CD63 Homologue from the Coleopteran Beetle, Tenebrio molitor. Int. J. Mol. Sci. 2013, 14, 20744-20767. https://doi.org/10.3390/ijms141020744
Patnaik BB, Kang SM, Seo GW, Lee HJ, Patnaik HH, Jo YH, Tindwa H, Lee YS, Lee BL, Kim NJ, et al. Molecular Cloning, Sequence Characterization and Expression Analysis of a CD63 Homologue from the Coleopteran Beetle, Tenebrio molitor. International Journal of Molecular Sciences. 2013; 14(10):20744-20767. https://doi.org/10.3390/ijms141020744
Chicago/Turabian StylePatnaik, Bharat Bhusan, Seong Min Kang, Gi Won Seo, Hyo Jeong Lee, Hongray Howrelia Patnaik, Yong Hun Jo, Hamisi Tindwa, Yong Seok Lee, Bok Luel Lee, Nam Jung Kim, and et al. 2013. "Molecular Cloning, Sequence Characterization and Expression Analysis of a CD63 Homologue from the Coleopteran Beetle, Tenebrio molitor" International Journal of Molecular Sciences 14, no. 10: 20744-20767. https://doi.org/10.3390/ijms141020744