Chromosome-Specific DNA Repeats: Rapid Identification in Silico and Validation Using Fluorescence in Situ Hybridization
Abstract
:1. Introduction
2. Results and Discussion
2.1. Hybridization Targets on the Human Gonosomes
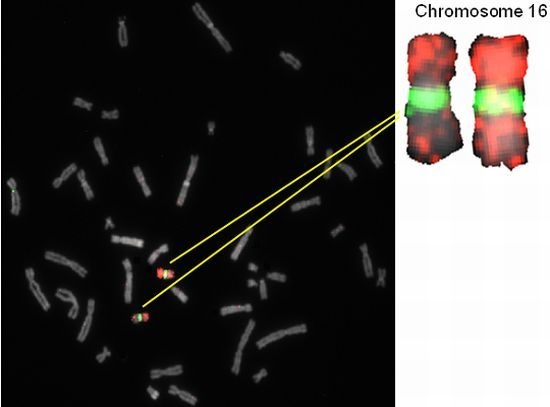
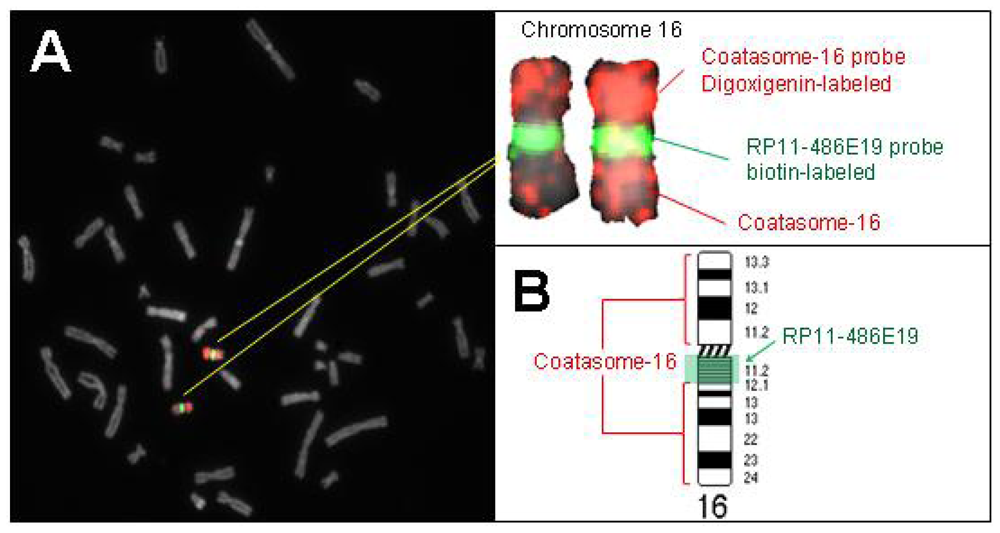
2.2. Hybridization Targets for Enumeration of Human Chromosome 16
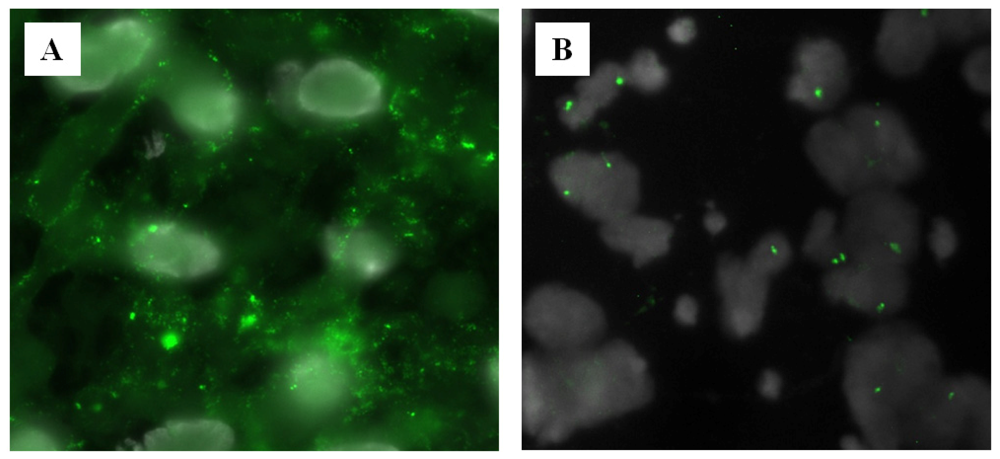
2.3. Clinical Perspective
3. Experimental Section
3.1. Choosing DNA Probes That Bind to Specific Human Chromosomes
3.1.1. Selection of Probes for Satellite-Rich Regions of Human Chromosomes 16 and X
3.1.2. Determining the Target Sequence on the Y Chromosome
3.1.3. Identifying a DNA Probe for the Target Sequence of Chromosome Y
3.2. Fluorescence in situ Hybridization
3.3. Pretreatment of Tissue Sections with RNase and Pepsin
3.4. Image Acquisition and Analysis
4. Conclusions
Acknowledgements
- DisclaimerThis document was prepared as an account of work sponsored by the United States Government. While this document is believed to contain correct information, neither the United States Government nor any agency thereof, nor The Regents of the University of California, nor any of their employees, makes any warranty, express or implied, or assumes any legal responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process, or service by its trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States Government or any agency thereof, or The Regents of the University of California. The views and opinions of authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof, or The Regents of the University of California.
- Conflict of InterestThe authors declare no conflict of interest.
References
- Weier, H.U.; Weier, J.F.; Renom, M.O.; Zheng, X.; Colls, P.; Nureddin, A.; Pham, C.D.; Chu, L.W.; Racowsky, C.; Munne, S. Fluorescence in situ hybridization and spectral imaging analysis of human oocytes and first polar bodies. J. Histochem. Cytochem 2005, 53, 269–272. [Google Scholar]
- Magli, M.C.; Gianaroli, L.; Crippa, A.; Munne, S.; Robles, F.; Ferraretti, A.P. Aneuploidies of chromosomes 1, 4, and 6 are not compatible with human embryos’ implantation. Fertil. Steril 2010, 94, 2012–2016. [Google Scholar]
- Kohn, G.; Ornoy, A.; Ben-Tsur, Z.; Sadovsky, E.; Cohen, M.M. Successive spontaneous abortions with diverse chromosomal aberrations in human translocation heterozygote. Teratology 1975, 12, 283–289. [Google Scholar]
- Hook, E.B.; Schreinemachers, D.M.; Willey, A.M.; Cross, P.K. Rates of mutant structural chromosome rearrangements in human fetuses: Data from prenatal cytogenetic studies and associations with maternal age and parental mutagen exposure. Am. J. Hum. Genet 1983, 35, 96–109. [Google Scholar]
- Callen, D.F.; Ringenbergs, M.L.; Fowler, J.C.; Freemantle, C.J.; Haan, E.A. Small marker chromosomes in man: Origin from pericentric heterochromatin of chromosomes 1, 9, and 16. J. Med. Genet 1990, 27, 155–159. [Google Scholar]
- Benadiva, C.A.; Kligman, I.; Munne, S. Aneuploidy 16 in human embryos increases significantly with maternal age. Fertil. Steril 1996, 66, 248–255. [Google Scholar]
- Munne, S.; Magli, C.; Bahce, M.; Fung, J.; Legator, M.; Morrison, L.; Cohert, J.; Gianaroli, L. Preimplantation diagnosis of the aneuploidies most commonly found in spontaneous abortions and live births: XY, 13, 14, 15, 16, 18, 21, 22. Prenat. Diagn 1998, 18, 1459–1466. [Google Scholar]
- Starke, H.; Nietzel, A.; Weise, A.; Heller, A.; Mrasek, K.; Belitz, B.; Kelbova, C.; Volleth, M.; Albrecht, B.; Mitulla, B.; et al. Small supernumerary marker chromosomes (SMCs): Genotype-phenotype correlation and classification. Hum. Genet 2003, 114, 51–67. [Google Scholar]
- Pflueger, S.M.V. Cytogenetics of spontaneous abortion. In The Principles of Clinical Cytogenetics, 2nd ed; Gersen, S.L., Keagle, M.B., Eds.; Humana Press Inc: Totowa, NJ, USA, 2005; pp. 323–345. [Google Scholar]
- Malmanche, N.; Maia, A.; Sunkel, C.E. The spindle assembly checkpoint: Preventing chromosome mis-segregation during mitosis and meiosis. FEBS Lett 2006, 580, 2888–2895. [Google Scholar]
- Colls, P.; Silver, L.; Olivera, G.; Weier, J.; Escudero, T.; Goodall, N.; Tomkin, G.; Munne, S. Preimplantation genetic diagnosis for gender selection in the USA. Reprod. Biomed. Online 2009, 19, 16–22. [Google Scholar]
- Hassold, T.J.; Jacobs, P.A. Trisomy in man. Annu. Rev. Genet 1984, 18, 69–97. [Google Scholar]
- Hassold, T.J. Chromosome abnormalities in human reproductive wastage. Trends Genet. 1986, 105–110. [Google Scholar]
- Lee, A.J.; Endesfelder, D.; Rowan, A.J.; Walther, A.; Birkbak, N.J.; Futreal, P.A.; Downward, J.; Szallasi, Z.; Tomlinson, I.P.; Howell, M.; et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res 2011, 71, 1858–1870. [Google Scholar]
- Lathi, R.B.; Westphal, L.M.; Milki, A.A. Aneuploidy in the miscarriages of infertile women and the potential benefit of preimplanation genetic diagnosis. Fertil. Steril 2008, 89, 353–357. [Google Scholar]
- Lichter, P.; Ward, D.C. Is non-isotopic in situ hybridization finally coming of age? Nature 1990, 345, 93–94. [Google Scholar]
- Weier, H.U.; Lucas, J.N.; Poggensee, M.; Segraves, R.; Pinkel, D.; Gray, J.W. Two-color hybridization with high complexity chromosome-specific probes and a degenerate alpha satellite probe DNA allows unambiguous discrimination between symmetrical and asymmetrical translocations. Chromosoma 1991, 100, 371–376. [Google Scholar]
- Weier, H.-U.; Pinkel, D.; Gray, J.W. Whole-chromosome complementary probe fluorescence staining. In Molecular Biology and Biotechnology; Meyers, R.A., Ed.; VCH Verlagsgesellschaft: Weinheim, Germany, 1995; pp. 965–968. [Google Scholar]
- Zitzelsberger, H.; Bruch, J.; Smida, J.; Hieber, L.; Peddie, C.M.; Bryant, P.E.; Riches, A.C.; Fung, J.; Weier, H.U.; Bauchinger, M. Clonal chromosomal aberrations in simian virus 40-transfected human thyroid cells and in derived tumors developed after in vitro irradiation. Int. J. Cancer 2001, 96, 166–177. [Google Scholar]
- Liehr, T.; Weise, A.; Heller, A.; Starke, H.; Mrasek, K.; Kuechler, A.; Weier, H.U.; Claussen, U. Multicolor chromosome banding (MCB) with YAC/BAC-based probes and region-specific microdissection DNA libraries. Cytogenet. Genome Res 2002, 97, 43–50. [Google Scholar]
- Zitzelsberger, H.; O’Brien, B.; Weier, H.U. Multicolor FISH techniques for the detection of inter- and intrachromosomal rearrangements. In FISH Technology; Rautenstrauss, B., Liehr, T., Eds.; Springer Verlag: Heidelberg, Germany, 2002; pp. 408–424. [Google Scholar]
- Liehr, T.; Nietzel, A.; Starke, H.; Heller, A.; Weise, A.; Kuechler, A.; Senger, G.; Ebner, S.; Martin, T.; Stumm, M.; et al. Characterization of small marker chromosomes (SMC) by recently developed molecular cytogenetic approaches. J. Assoc. Genet. Technol 2003, 29, 5–10. [Google Scholar]
- O’Brien, B.; Jossart, G.H.; Ito, Y.; Greulich-Bode, K.M.; Weier, J.F.; Munne, S.; Clark, O.H.; Weier, H.U.G. Chromosomal Rainbows’ detect oncogenic rearrangements of signaling molecules in thyroid tumors. Open Cell Signal. J 2010, 2, 13–21. [Google Scholar]
- Jossart, G.H.; O’Brien, B.; Cheng, J.F.; Tong, Q.; Jhiang, S.M.; Duh, Q.; Clark, O.H.; Weier, H.U. A novel multicolor hybridization scheme applied to localization of a transcribed sequence (D10S170/H4) and deletion mapping in the thyroid cancer cell line TPC-1. Cytogenet. Cell Genet 1996, 75, 254–257. [Google Scholar]
- Cassel, M.J.; Munne, S.; Fung, J.; Weier, H.U. Carrier-specific breakpoint-spanning DNA probes: An approach to preimplantation genetic diagnosis in interphase cells. Hum. Reprod 1997, 12, 2019–2027. [Google Scholar]
- Weier, H.U.; Rhein, A.P.; Shadravan, F.; Collins, C.; Polikoff, D. Rapid physical mapping of the human trk protooncogene (NTRK1) to human chromosome 1q21-q22 by P1 clone selection, fluorescence in situ hybridization (FISH), and computer-assisted microscopy. Genomics 1995, 26, 390–393. [Google Scholar]
- Greulich, K.M.; Kreja, L.; Heinze, B.; Rhein, A.P.; Weier, H.G.; Bruckner, M.; Fuchs, P.; Molls, M. Rapid detection of radiation-induced chromosomal aberrations in lymphocytes and hematopoietic progenitor cells by mFISH. Mutat. Res 2000, 452, 73–81. [Google Scholar]
- Greulich-Bode, K.M.; Wang, M.; Rhein, A.P.; Weier, J.F.; Weier, H.U. Validation of DNA probes for molecular cytogenetics by mapping onto immobilized circular DNA. Mol. Cytogenet 2008, 1, 28. [Google Scholar]
- Chen, X.N.; Korenberg, J.R. BAC resource for molecular cytogenetics. Methods Mol. Biol 2002, 204, 391–403. [Google Scholar]
- Munne, S.; Howles, C.M.; Wells, D. The role of preimplantation genetic diagnosis in diagnosing embryo aneuploidy. Curr. Opin. Obstet. Gynecol 2009, 21, 442–449. [Google Scholar]
- Weier, H.U.; Segraves, R.; Pinkel, D.; Gray, J.W. Synthesis of Y chromosome-specific labeled DNA probes by in vitro DNA amplification. J. Histochem. Cytochem 1990, 38, 421–426. [Google Scholar]
- Munne, S.; Grifo, J.; Cohen, J.; Weier, H.U. Chromosome abnormalities in human arrested preimplantation embryos: A multiple-probe FISH study. Am. J. Hum. Genet 1994, 55, 150–159. [Google Scholar]
- Weier, H.U.; Polikoff, D.; Fawcett, J.J.; Greulich, K.M.; Lee, K.H.; Cram, S.; Chapman, V.M.; Gray, J.W. Generation of five high-complexity painting probe libraries from flow-sorted mouse chromosomes. Genomics 1994, 21, 641–644. [Google Scholar]
- Munne, S.; Sultan, K.M.; Weier, H.U.; Grifo, J.A.; Cohen, J.; Rosenwaks, Z. Assessment of numeric abnormalities of X, Y, 18, and 16 chromosomes in preimplantation human embryos before transfer. Am. J. Obstet. Gynecol 1995, 172, 1191–1199. [Google Scholar]
- Munne, S.; Weier, H.U. Simultaneous enumeration of chromosomes 13, 18, 21, X, and Y in interphase cells for preimplantation genetic diagnosis of aneuploidy. Cytogenet. Cell Genet 1996, 75, 263–270. [Google Scholar]
- Weier, H.U.; Tuton, T.B.; Ito, Y.; Chu, L.W.; Lu, C.M.; Baumgartner, A.; Zitzelsberger, H.F.; Weier, J.F. Molecular cytogenetic characterization of chromosome 9-derived material in a human thyroid cancer cell line. Cytogenet. Genome Res 2006, 114, 284–291. [Google Scholar]
- Lu, C.M.; Kwan, J.; Baumgartner, A.; Weier, J.F.; Wang, M.; Escudero, T.; Munne, S.; Zitzelsberger, H.F.; Weier, H.U. DNA probe pooling for rapid delineation of chromosomal breakpoints. J. Histochem. Cytochem 2009, 57, 587–597. [Google Scholar]
- Zeng, H.; Weier, H.U.G.; Kwan, J.; Wang, M.; O’Brien, B. Data mining empowers the generation of a novel class of chromosome-specific DNA probes. J. Data Min. Genomics Proteomics 2011, 2, 108. [Google Scholar]
- Christensen, B.; Bryndorf, T.; Philip, J.; Lundsteen, C.; Hansen, W. Rapid prenatal diagnosis of trisomy 18 and triploidy in interphase nuclei of uncultured amniocytes by non-radioactive in situ hybridization. Prenat. Diagn 1992, 12, 241–250. [Google Scholar]
- Waye, J.S.; Durfy, S.J.; Pinkel, D.; Kenwrick, S.; Patterson, M.; Davies, K.E.; Willard, H.F. Chromosome-specific alpha satellite DNA from human chromosome 1: Hierarchical structure and genomic organization of a polymorphic domain spanning several hundred kilobase pairs of centromeric DNA. Genomics 1987, 1, 43–51. [Google Scholar]
- Waye, J.S.; Willard, H.F. Concerted evolution of alpha satellite DNA: Evidence for species specificity and a general lack of sequence conservation among alphoid sequences of higher primates. Chromosoma 1989, 98, 273–279. [Google Scholar]
- Weier, H.U.; Kleine, H.D.; Gray, J.W. Labeling of the centromeric region on human chromosome 8 by in situ hybridization. Hum. Genet 1991, 87, 489–494. [Google Scholar]
- Weier, H.U.; Gray, J.W. A degenerate alpha satellite probe, detecting a centromeric deletion on chromosome 21 in an apparently normal human male, shows limitations of the use of satellite DNA probes for interphase ploidy analysis. Anal. Cell. Pathol 1992, 4, 81–86. [Google Scholar]
- Baumgartner, A.; Weier, J.F.; Weier, H.U. Chromosome-specific DNA repeat probes. J. Histochem. Cytochem 2006, 54, 1363–1370. [Google Scholar]
- Lu, C.M.; Kwan, J.; Weier, J.F.; Baumgartner, A.; Wang, M.; Escudero, T.; Munne, S.; Weier, H.U. Rapid mapping of chromosomal breakpoints: from blood to BAC in 20 days. Folia Histochem. Cytobiol 2009, 47, 367–375. [Google Scholar]
- Kalousek, D.K. Pathogenesis of chromosomal mosaicism and its effect on early human development. Am. J. Med. Genet 2000, 91, 39–45. [Google Scholar]
- Guze, C. Human genetics-chromosome abnormalities. Carol’s Classroom Website. Available online: http://www.carolguze.com/text/442-5-chromosome_abnormalities.shtml accessed on 13 April 2011.
- Philip, J.; Bryndorf, T.; Christensen, B. Prenatal aneuploidy detection in interphase cells by fluorescence in situ hybridization (FISH). Prenat. Diagn 1994, 14, 1203–1215. [Google Scholar]
- Bryndorf, T. Development and clinical studies of in situ hybridization techniques in prenatal diagnosis. Dan. Med. Bull 1998, 45, 298–312. [Google Scholar]
- Bryndorf, T.; Lundsteen, C.; Lamb, A.; Christensen, B.; Philip, J. Rapid prenatal diagnosis of chromosome aneuploidies by interphase fluorescence in situ hybridization: A one-year clinical experience with high-risk and urgent fetal and postnatal samples. Acta Obstet. Gynecol. Scand 2000, 79, 8–14. [Google Scholar]
- Tarailo-Graovac, M.; Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics 2009, 25. [Google Scholar] [CrossRef]
- Nakahori, Y.; Mitani, K.; Yamada, M.; Nakagome, Y. A human Y-chromosome specific repeated DNA family (DYZ1) consists of a tandem array of pentanucleotides. Nucleic Acids Res 1986, 14, 7569–7580. [Google Scholar]
- Wolstenholme, J. An audit of trisomy 16 in man. Prenat. Diagn 1995, 15, 109–121. [Google Scholar]
- Gardner, R.J.M.; Sutherland, G.R. Chromosome Abnormalities and Genetic Counseling, 2nd ed; Oxford University Press: New York, NY, USA, 1996; pp. 59–190. [Google Scholar]
- Ancel, P.Y.; Saurel-Cubizolles, M.J.; Di Renzo, G.C.; Papiernik, E.; Breart, G. Risk factors for 14–21 week abortions: A case-control study in Europe. The Europop Group. Hum. Reprod 2000, 15, 2426–2432. [Google Scholar]
- Delhanty, J.D.; Harper, J.C.; Ao, A.; Handyside, A.H.; Winston, R.M. Multicolour FISH detects frequent chromosomal mosaicism and chaotic division in normal preimplantation embryos from fertile patients. Hum. Genet 1997, 99, 755–760. [Google Scholar]
- Weier, H.U.; Munne, S.; Fung, J. Patient-specific probes for preimplantation genetic diagnosis of structural and numerical aberrations in interphase cells. J. Assist. Reprod. Genet 1999, 16, 182–191. [Google Scholar]
- Verlinsky, Y.; Tur-Kaspa, I.; Cieslak, J.; Bernal, A.; Morris, R.; Taranissi, M.; Kaplan, B.; Kuliev, A. Preimplantation testing for chromosomal disorders improves reproductive outcome of poor-prognosis patients. Reprod. Biomed. Online 2005, 11, 219–225. [Google Scholar]
- Munné, S. Preimplantation genetic diagnosis for aneuploidy and translocations using array comparative genomic hybridization. Curr. Genomics 2012, 13, 463–470. [Google Scholar]
- Practice Committee of Society for Assisted Reproductive Technology and Practice Committee of American Society for Reproductive Medicine. Preimplantation genetic testing: A Practice Committee opinion. Fertil. Steril. 2007, 88, 1497–1504.
- Soini, S.; Ibarreta, D.; Anastasiadou, V.; Ayme, S.; Braga, S.; Cornel, M.; Coviello, D.A.; Evers-Kiebooms, G.; Geraedts, J.; Gianaroli, L.; et al. The interface between assisted reproductive technologies and genetics: Technical, social, ethical and legal issues. Eur. J. Hum. Genet 2006, 14, 588–645. [Google Scholar]
- Otani, T.; Roche, M.; Mizuike, M.; Colls, P.; Escudero, T.; Munne, S. Preimplantation genetic diagnosis significantly improves the pregnancy outcome of translocation carriers with a history of recurrent miscarriage and unsuccessful pregnancies. Reprod. Biomed. Online 2006, 13, 869–874. [Google Scholar]
- Fiorentino, F.; Kokkali, G.; Biricik, A.; Stavrou, D.; Ismailoglu, B.; de Palma, R.; Arizzi, L.; Harton, G.; Sessa, M.; Pantos, K. Polymerase chain reaction-based detection of chromosomal imbalances on embryos: The evolution of preimplantation genetic diagnosis for chromosomal translocations. Fertil. Steril 2010, 94, 2001–2011. [Google Scholar]
- Cooke, H.J.; Hindley, J. Cloning of human satellite III DNA: Different components are on different chromosomes. Nucleic Acids Res 1979, 6, 3177–3197. [Google Scholar]
- Cooke, H.J.; McKay, R.D. Evolution of a human Y chromosome-specific repeated sequence. Cell 1978, 13, 453–460. [Google Scholar]
- Weier, H.U.; Reitsma, M.; Gray, J.W. Detection of fetal cells by in vitro DNA amplification. Analyt. Cell. Pathol 1989, 1, 313. [Google Scholar]
- Weier, H.U.; Reitsma, M.; Gray, J.W. Detection of fetal cells by in vitro DNA amplification. In Advances in Analytical Cellular Pathology; Proceedings of the First Conference of the European Society for Analytical Cellular Pathology, Schloss Elmau, F.R.G. Burger, G., Oberholzer, M., Vooijs, G.P., Eds.; Elsevier Science Publishers: Amsterdam, The Netherlands, 1990; pp. 105–106. [Google Scholar]
- Gray, J.W.; Weier, H.-U. Y chromosome specific nucleic acid probe and method for determining the Y in situ. U.S. Patent 6300066, 9 October 2001. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol 1990, 215, 403–410. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 1997, 25, 3389–3402. [Google Scholar]
- NCBI/BLAST Home Page. Available online: http://ncbi.nih.gov/blast accessed on 14 December 2012.
- Weier, H.U. Quantitative DNA fiber mapping. Methods Cell Biol 2001, 64, 33–53. [Google Scholar]
- Weier, H.U.; Wang, M.; Mullikin, J.C.; Zhu, Y.; Cheng, J.F.; Greulich, K.M.; Bensimon, A.; Gray, J.W. Quantitative DNA fiber mapping. Hum. Mol. Genet 1995, 4, 1903–1910. [Google Scholar]
- Fung, J.; Hyun, W.; Dandekar, P.; Pedersen, R.A.; Weier, H.U. Spectral imaging in preconception/preimplantation genetic diagnosis of aneuploidy: Multicolor, multichromosome screening of single cells. J. Assist. Reprod. Genet 1998, 15, 323–330. [Google Scholar]
- Fung, J.; Weier, H.U.; Pedersen, R.A.; Zitzelsberger, H. Spectral imaging analysis of metaphase and interphase cells. In FISH Technology; Rautenstrauss, B., Liehr, T., Eds.; Springer Verlag: Heidelberg, Germany, 2002; pp. 363–387. [Google Scholar]
- Kwan, J.; Baumgartner, A.; Lu, C.M.; Wang, M.; Weier, J.F.; Zitzelsberger, H.F.; Weier, H.U. BAC-FISH assays delineate complex chromosomal rearrangements in a case of post-Chernobyl childhood thyroid cancer. Folia Histochem. Cytobiol 2009, 47, 135–142. [Google Scholar]
- Kalousek, D.K.; Vekemans, M. Confined placental mosaicism. J. Med. Genet 1996, 33, 529–533. [Google Scholar]
- Benn, P. Trisomy 16 and trisomy 16 Mosaicism: A review. Am. J. Med. Genet 1998, 79, 121–133. [Google Scholar]
- Yong, P.J.; Barrett, I.J.; Kalousek, D.K.; Robinson, W.P. Clinical aspects, prenatal diagnosis, and pathogenesis of trisomy 16 mosaicism. J. Med. Genet 2003, 40, 175–182. [Google Scholar]
- Fung, J.; Munné, S.; Duell, T.; Weier, H.-U.G. Rapid cloning of translocation breakpoints: From blood to YAC in 50 days. J. Biochem. Mol. Biol. Biophys 1998, 1, 181–192. [Google Scholar]



| BAC | Chr. | Band | Start (bp) * | End (bp) * | Insert (bp) | BAC End Sequence Accession Number |
|---|---|---|---|---|---|---|
| RP11-242E13 | Y | q12 | multiple | multiple | 98295 | AC068123 |
| RP11-348G24 | X | p11.1 | 58,356,061 | 58,564,667 | 208607 | AQ528470, AQ528473 |
| RP11-88G23 | 16 | q11.2 | 46,385,822 | 46,412,445 | 26624 | AQ285754, AQ285753 |
| RP11-246K16 | 16 | q11.2 | 46,385,808 | 46,412,485 | 26678 | AQ478385, AQ478388 |
| RP11-416F8 | 16 | q11.2 | 46,385,808 | 46,412,485 | 26678 | AQ551230, AQ661227 |
| RP11-486E19 | 16 | q11.2 | 46,385,921 | 46,412,470 | 26550 | AQ629869, AQ629871 |
© 2013 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hsu, J.H.; Zeng, H.; Lemke, K.H.; Polyzos, A.A.; Weier, J.F.; Wang, M.; Lawin-O'Brien, A.R.; Weier, H.-U.G.; O'Brien, B. Chromosome-Specific DNA Repeats: Rapid Identification in Silico and Validation Using Fluorescence in Situ Hybridization. Int. J. Mol. Sci. 2013, 14, 57-71. https://doi.org/10.3390/ijms14010057
Hsu JH, Zeng H, Lemke KH, Polyzos AA, Weier JF, Wang M, Lawin-O'Brien AR, Weier H-UG, O'Brien B. Chromosome-Specific DNA Repeats: Rapid Identification in Silico and Validation Using Fluorescence in Situ Hybridization. International Journal of Molecular Sciences. 2013; 14(1):57-71. https://doi.org/10.3390/ijms14010057
Chicago/Turabian StyleHsu, Joanne H., Hui Zeng, Kalistyn H. Lemke, Aris A. Polyzos, Jingly F. Weier, Mei Wang, Anna R. Lawin-O'Brien, Heinz-Ulrich G. Weier, and Benjamin O'Brien. 2013. "Chromosome-Specific DNA Repeats: Rapid Identification in Silico and Validation Using Fluorescence in Situ Hybridization" International Journal of Molecular Sciences 14, no. 1: 57-71. https://doi.org/10.3390/ijms14010057
APA StyleHsu, J. H., Zeng, H., Lemke, K. H., Polyzos, A. A., Weier, J. F., Wang, M., Lawin-O'Brien, A. R., Weier, H.-U. G., & O'Brien, B. (2013). Chromosome-Specific DNA Repeats: Rapid Identification in Silico and Validation Using Fluorescence in Situ Hybridization. International Journal of Molecular Sciences, 14(1), 57-71. https://doi.org/10.3390/ijms14010057