Direct Observation of Protein Microcrystals in Crystallization Buffer by Atmospheric Scanning Electron Microscopy
Abstract
:1. Introduction
2. Results and Discussion
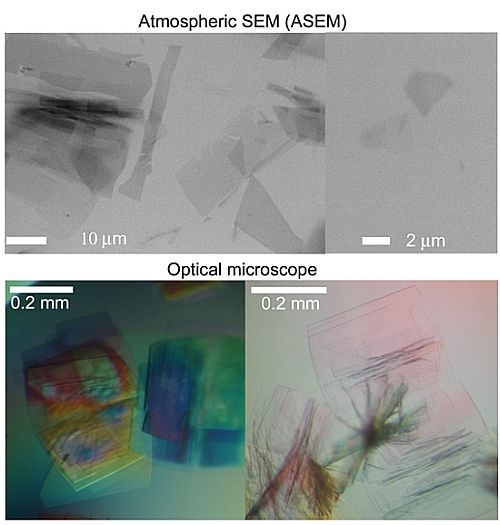
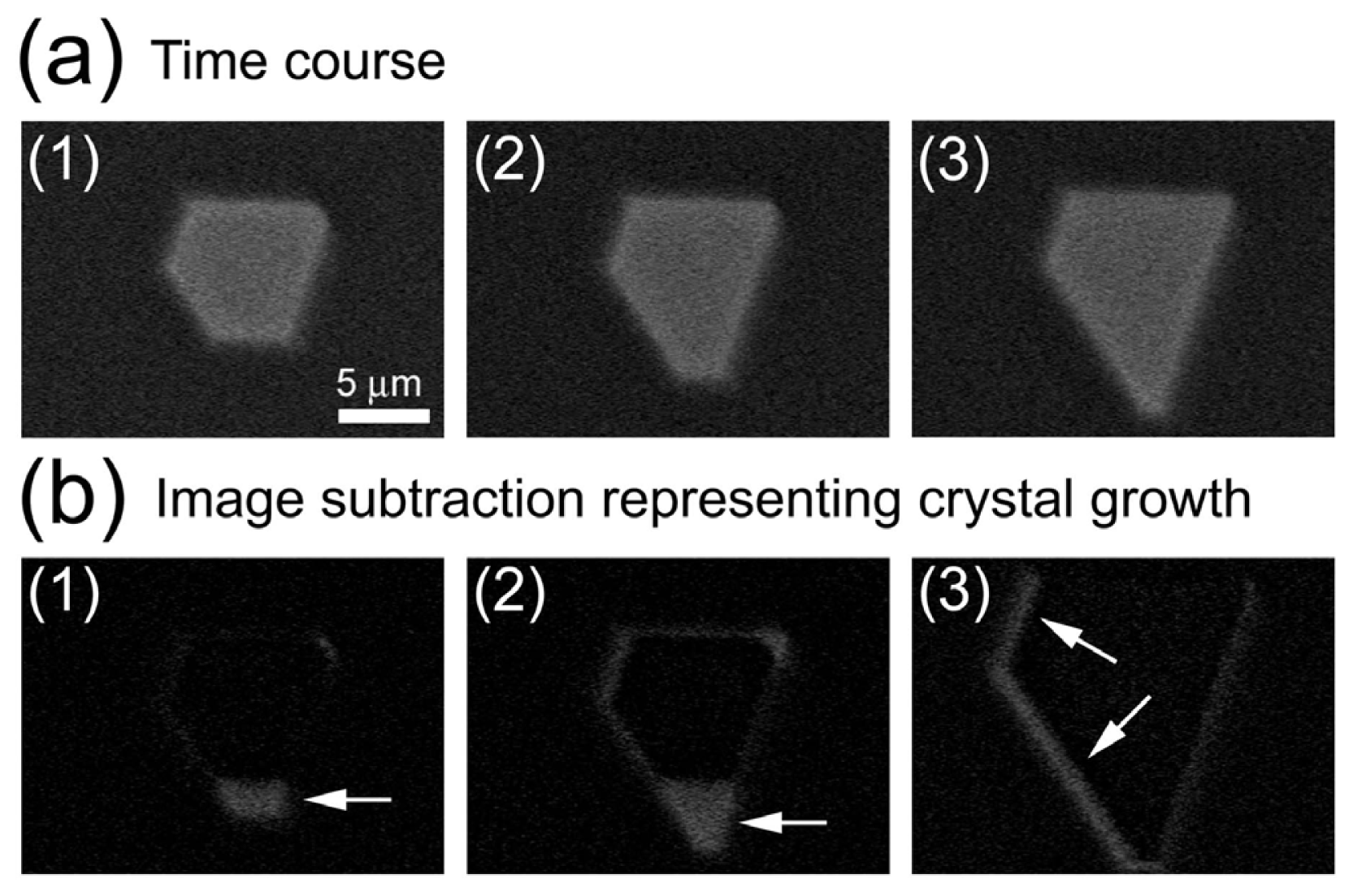
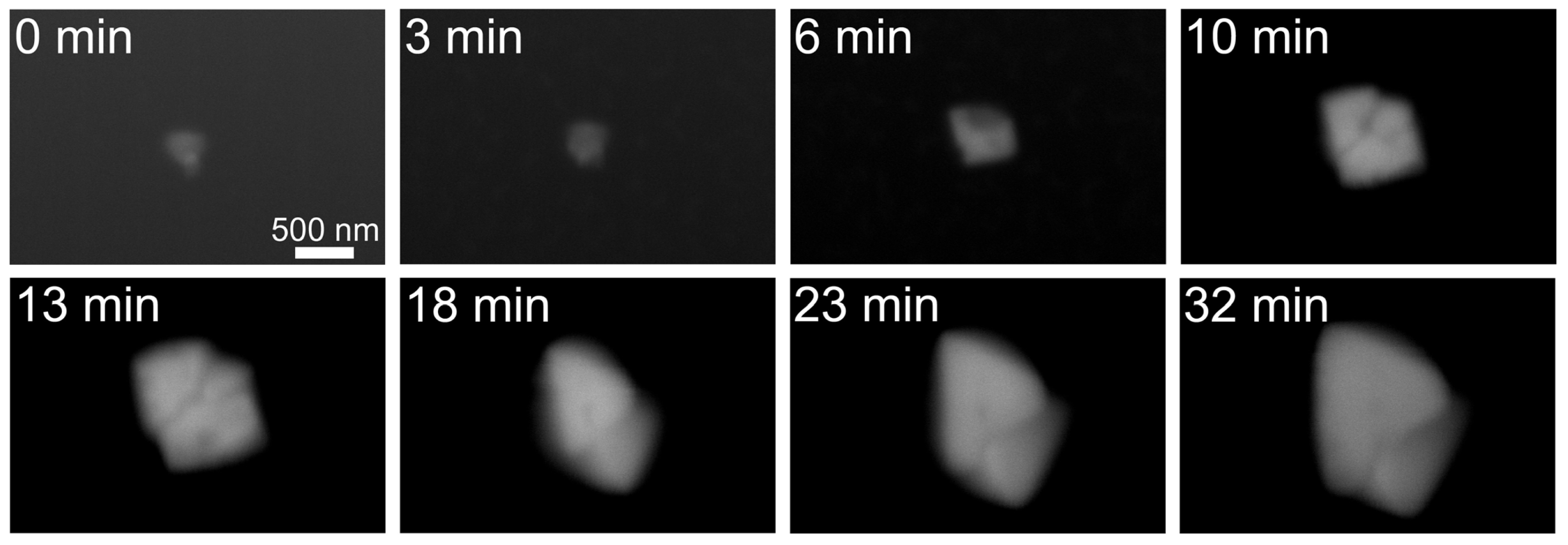
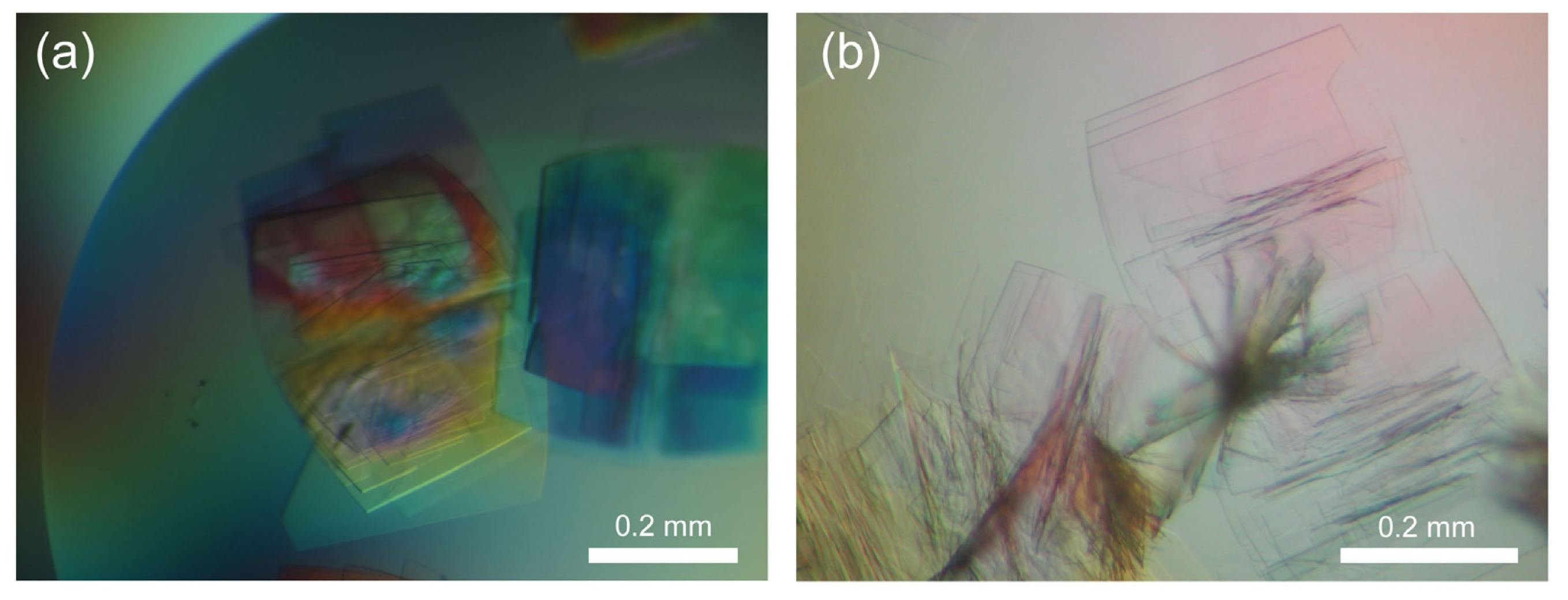
2.1. Observation of Salt Crystals and Their Growth
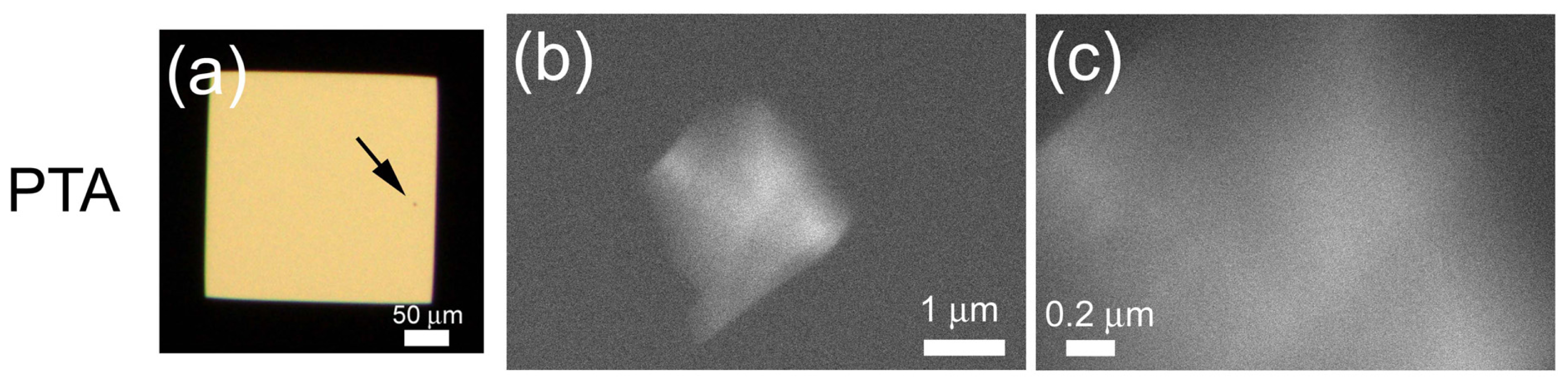
2.2. Metal Staining of Protein Crystals
2.3. Staining of Lysozyme Micro-Crystals
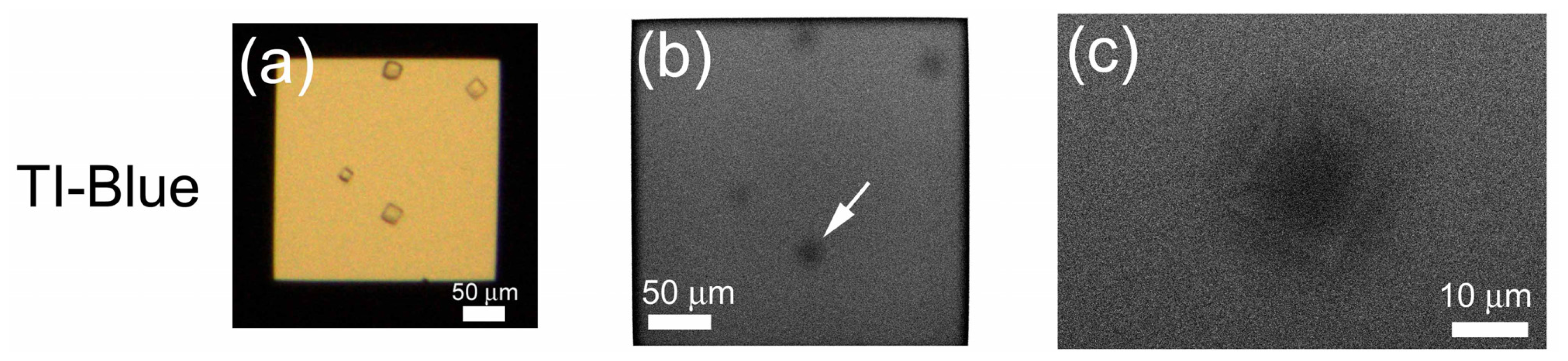
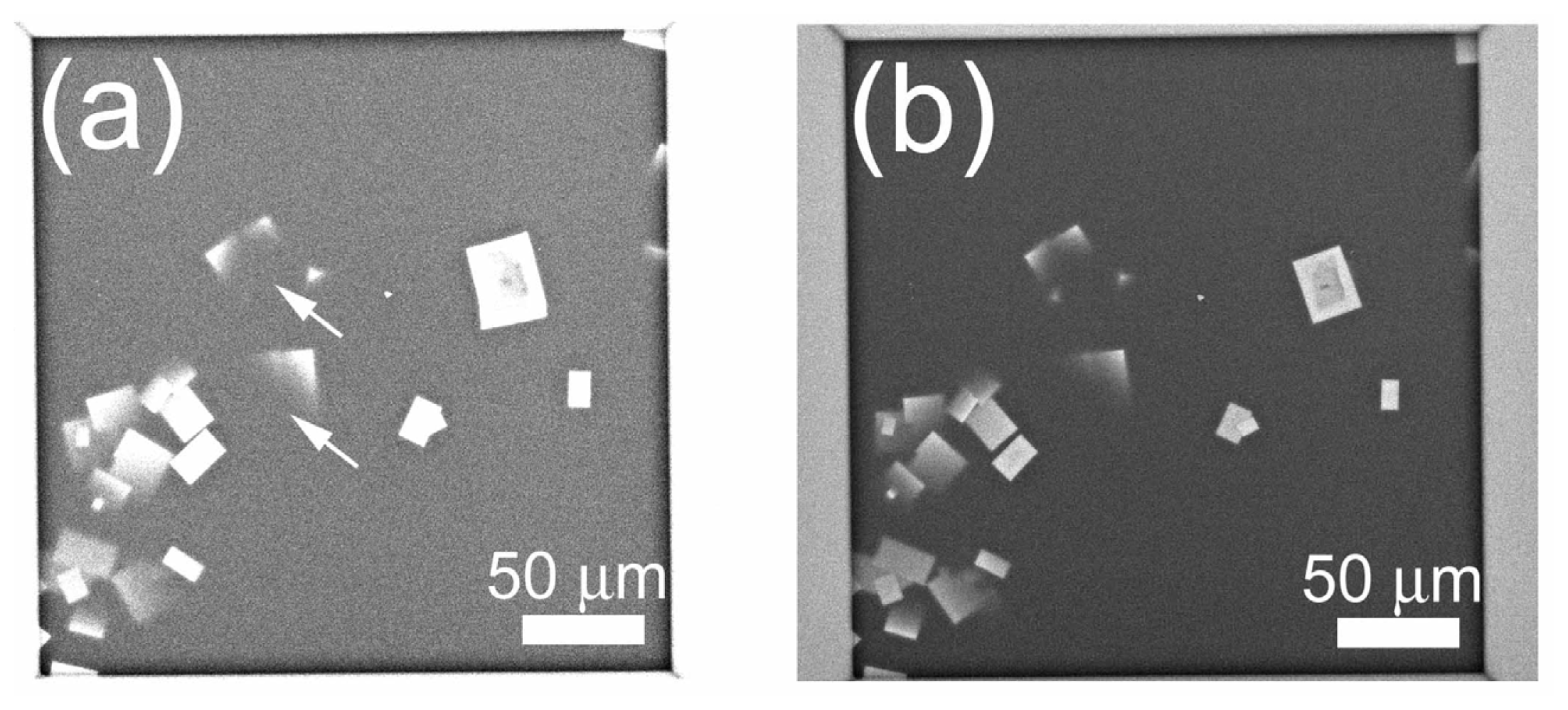
2.4. Non-Stained Lysozyme Protein Crystal in Crystallization Buffer
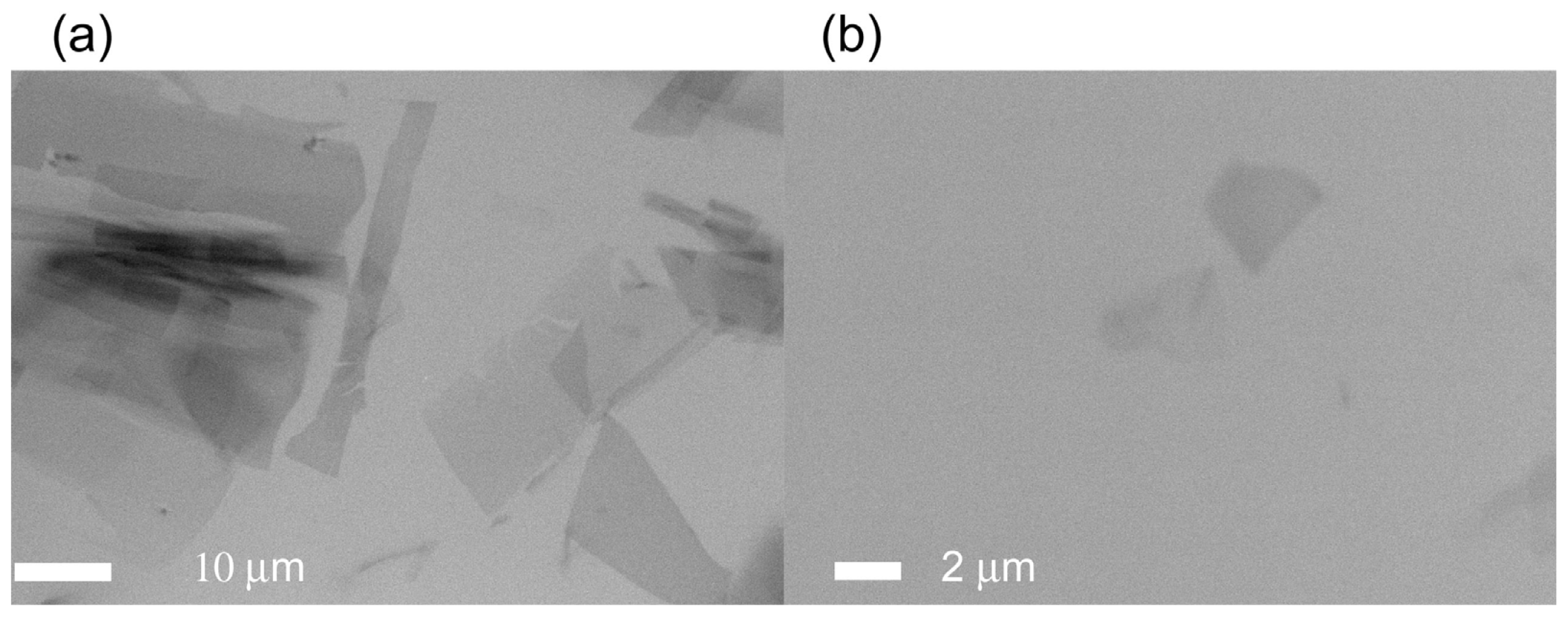
2.5. Observation of Non-Stained TAF-Iβ Crystal
2.6. In Situ Observation of Non-Stained Lipase B Crystal
2.7. Difference between Salt and Protein Crystals
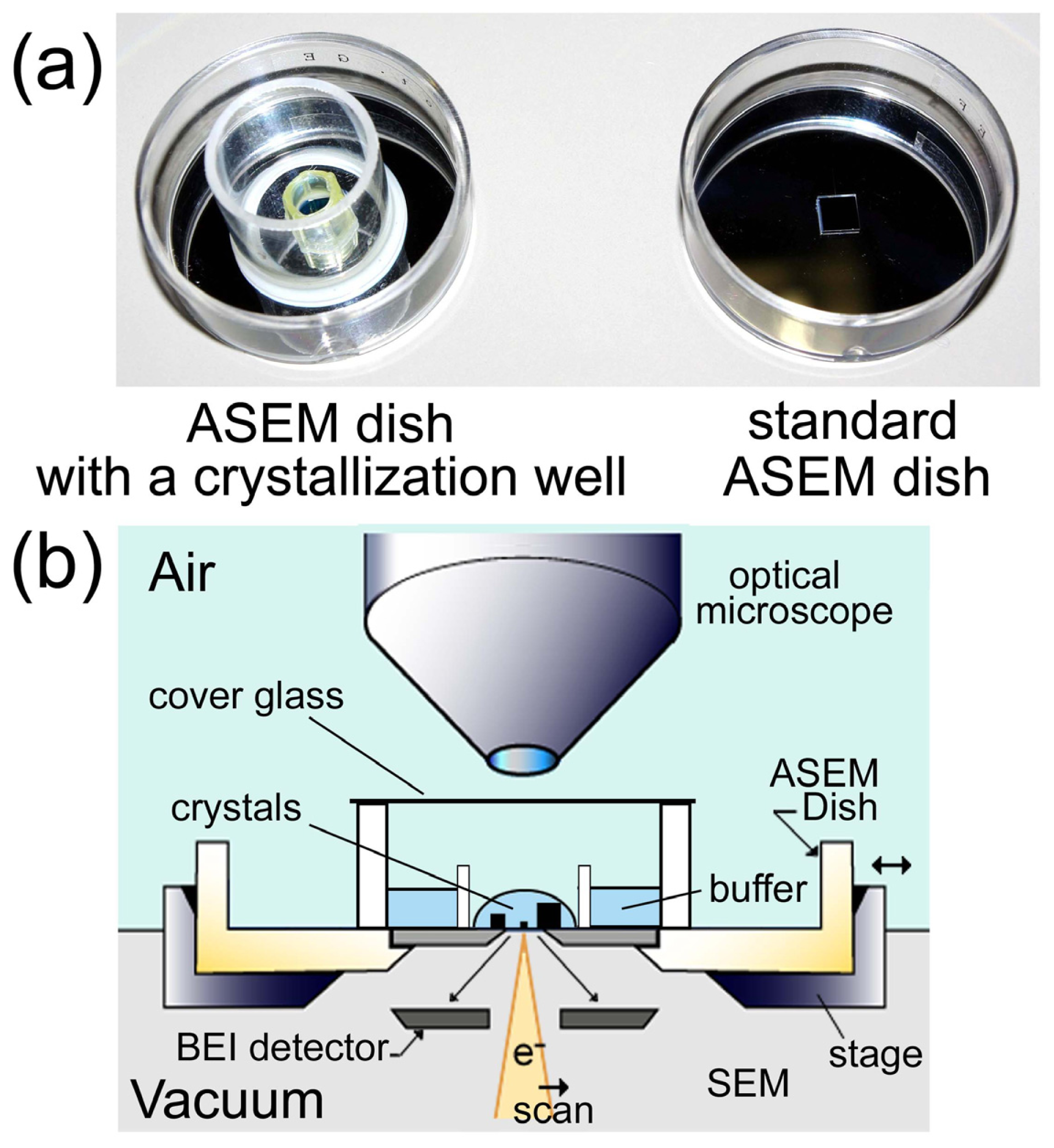
2.8. ASEM Dishes for Protein Crystallization
2.9. Comparison of ASEM with Other High-Resolution Microscopy
3. Experimental Section
3.1. Crystallization
3.2. Heavy Metal Staining of Protein Crystals
3.3. Imaging
4. Conclusions
Acknowledgments
- Conflict of InterestThe authors declare no conflict of interest.
References
- Branden, C.; Tooze, J. Introduction to Protein Structure; Garland Science: New York, NY, USA; p. 1999.
- Kato, H.E.; Zhang, F.; Yizhar, O.; Ramakrishnan, C.; Nishizawa, T.; Hirata, K.; Ito, J.; Aita, Y.; Tsukazaki, T.; Hayashi, S.; et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature 2012, 482, 369–374. [Google Scholar]
- Maeda, S.; Nakagawa, S.; Suga, M.; Yamashita, E.; Oshima, A.; Fujiyoshi, Y.; Tsukihara, T. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature 2009, 458, 597–602. [Google Scholar]
- Wüthrich, K. NMR of Proteins and Nucleic Acids; Wiley-Interscience: Hoboken, NJ, USA; p. 1986.
- Frank, J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies: Visualization of Biological Molecules in Their Native State; Oxford University Press: New York, NY, USA, 2006. [Google Scholar]
- Ogura, T.; Tong, K.I.; Mio, K.; Maruyama, Y.; Kurokawa, H.; Sato, C.; Yamamoto, M. Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening, double glycine repeat, and C-terminal domains. Proc. Natl. Acad. Sci. USA 2010, 107, 2842–2847. [Google Scholar]
- Sato, C.; Ueno, Y.; Asai, K.; Takahashi, K.; Sato, M.; Engel, A.; Fujiyoshi, Y. The voltage-sensitive sodium channel is a bell-shaped molecule with several cavities. Nature 2001, 409, 1047–1051. [Google Scholar]
- Riekel, C.; Burghammer, M.; Schertler, G. Protein crystallography microdiffraction. Curr. Opin. Struct. Biol 2005, 15, 556–562. [Google Scholar]
- Coulibaly, F.; Chiu, E.; Ikeda, K.; Gutmann, S.; Haebel, P.W.; Schulze-Briese, C.; Mori, H.; Metcalf, P. The molecular organization of cypovirus polyhedra. Nature 2007, 446, 97–101. [Google Scholar]
- Chapman, H.N.; Fromme, P.; Barty, A.; White, T.A.; Kirian, R.A.; Aquila, A.; Hunter, M.S.; Schulz, J.; DePonte, D.P.; Weierstall, U.; et al. Femtosecond X-ray protein nanocrystallography. Nature 2011, 470, 73–77. [Google Scholar]
- Bachmann, L.; Becker, R.; Leupold, G.; Barth, M.; Guckenberger, R.; Baumeister, W. Decoration and shadowing of freeze-etched catalase crystals. Ultramicroscopy 1985, 16, 305–320. [Google Scholar]
- Durbin, S.D.; Feher, G. Studies of crystal growth mechanisms of proteins by electron microscopy. J. Mol. Biol 1990, 212, 763–774. [Google Scholar]
- De Jonge, N.; Peckys, D.B.; Kremers, G.J.; Piston, D.W. Electron microscopy of whole cells in liquid with nanometer resolution. Proc. Natl. Acad. Sci. USA 2009, 106, 2159–2164. [Google Scholar]
- Zheng, H.; Smith, R.K.; Jun, Y.W.; Kisielowski, C.; Dahmen, U.; Alivisatos, A.P. Observation of single colloidal platinum nanocrystal growth trajectories. Science 2009, 324, 1309–1312. [Google Scholar]
- Nishiyama, H.; Suga, M.; Ogura, T.; Maruyama, Y.; Koizumi, M.; Mio, K.; Kitamura, S.; Sato, C. Atmospheric scanning electron microscope observes cells and tissues in open medium through silicon nitride film. J. Struct. Biol 2010, 169, 438–449. [Google Scholar]
- Suga, M.; Nishiyama, H.; Konyuba, Y.; Iwamatsu, S.; Watanabe, Y.; Yoshiura, C.; Ueda, T.; Sato, C. The atmospheric scanning electron microscope with open sample space observes dynamic phenomena in liquid or gas. Ultramicroscopy 2011, 111, 1650–1658. [Google Scholar]
- Mullin, J.W. Crystallization, 4th ed; Butterworth-Heinemann: Oxford, UK; p. 2001.
- Li, M.; Nadarajah, A.; Pusey, M.L. Growth of (101) faces of tetragonal lysozyme crystals: Determination of the growth mechanism. Acta Crystallogr. Sect. Biol. Crystallogr 1999, 55, 1012–1022. [Google Scholar]
- Muto, S.; Senda, M.; Adachi, N.; Suzuki, T.; Nagai, R.; Senda, T.; Horikoshi, M. Purification, crystallization and preliminary X-ray diffraction analysis of human oncoprotein SET/TAF-1beta. Acta Crystallogr. Sect. Biol. Crystallogr 2004, 60, 712–714. [Google Scholar]
- Judge, R.A.; Swift, K.; Gonzalez, C. An ultraviolet fluorescence-based method for identifying and distinguishing protein crystals. Acta Crystallogr. Sect. Biol. Crystallogr 2005, 61, 60–66. [Google Scholar]
- Dierks, K.; Meyer, A.; Einspahr, H.; Betzel, C. Dynamic light scattering in protein crystallization droplets: Adaptations for analysis and optimization of crystallization processes. Cryst. Growth Des 2008, 8, 1628–1634. [Google Scholar]
- Kissick, D.J.; Gualtieri, E.J.; Simpson, G.J. Nonlinear optical imaging of integral membrane protein crystals in lipidic mesophases. Anal. Chem 2010, 82, 491–497. [Google Scholar]
- Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol 1968, 33, 491–497. [Google Scholar]
- Thiberge, S.; Nechushtan, A.; Sprinzak, D.; Gileadi, O.; Behar, V.; Zik, O.; Chowers, Y.; Michaeli, S.; Schlessinger, J.; Moses, E. Scanning electron microscopy of cells and tissues under fully hydrated conditions. Proc. Natl. Acad. Sci. USA 2004, 101, 3346–3351. [Google Scholar]
- Engel, A. Biological applications of scanning probe microscopes. Annu. Rev. Biophys. Biophys. Chem 1991, 20, 79–108. [Google Scholar]
- Durbin, S.D.; Carlson, W.E. Lysozyme crystal-growth studied by atomic force microscopy. J. Cryst. Growth 1992, 122, 71–79. [Google Scholar]
- Durbin, S.D.; Carlson, W.E.; Saros, M.T. In situ studies of protein crystal growth by atomic force microscopy. J. Phys. Appl. Phys 1993, 26, B128–B132. [Google Scholar]




© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Maruyama, Y.; Ebihara, T.; Nishiyama, H.; Konyuba, Y.; Senda, M.; Numaga-Tomita, T.; Senda, T.; Suga, M.; Sato, C. Direct Observation of Protein Microcrystals in Crystallization Buffer by Atmospheric Scanning Electron Microscopy. Int. J. Mol. Sci. 2012, 13, 10553-10567. https://doi.org/10.3390/ijms130810553
Maruyama Y, Ebihara T, Nishiyama H, Konyuba Y, Senda M, Numaga-Tomita T, Senda T, Suga M, Sato C. Direct Observation of Protein Microcrystals in Crystallization Buffer by Atmospheric Scanning Electron Microscopy. International Journal of Molecular Sciences. 2012; 13(8):10553-10567. https://doi.org/10.3390/ijms130810553
Chicago/Turabian StyleMaruyama, Yuusuke, Tatsuhiko Ebihara, Hidetoshi Nishiyama, Yuji Konyuba, Miki Senda, Takuro Numaga-Tomita, Toshiya Senda, Mitsuo Suga, and Chikara Sato. 2012. "Direct Observation of Protein Microcrystals in Crystallization Buffer by Atmospheric Scanning Electron Microscopy" International Journal of Molecular Sciences 13, no. 8: 10553-10567. https://doi.org/10.3390/ijms130810553