Elongation Factor 1β' Gene from Spodoptera exigua: Characterization and Function Identification through RNA Interference
Abstract
:1. Introduction
2. Results and Discussion
2.1. Sequence Analysis of SeEF-1β′ cDNA
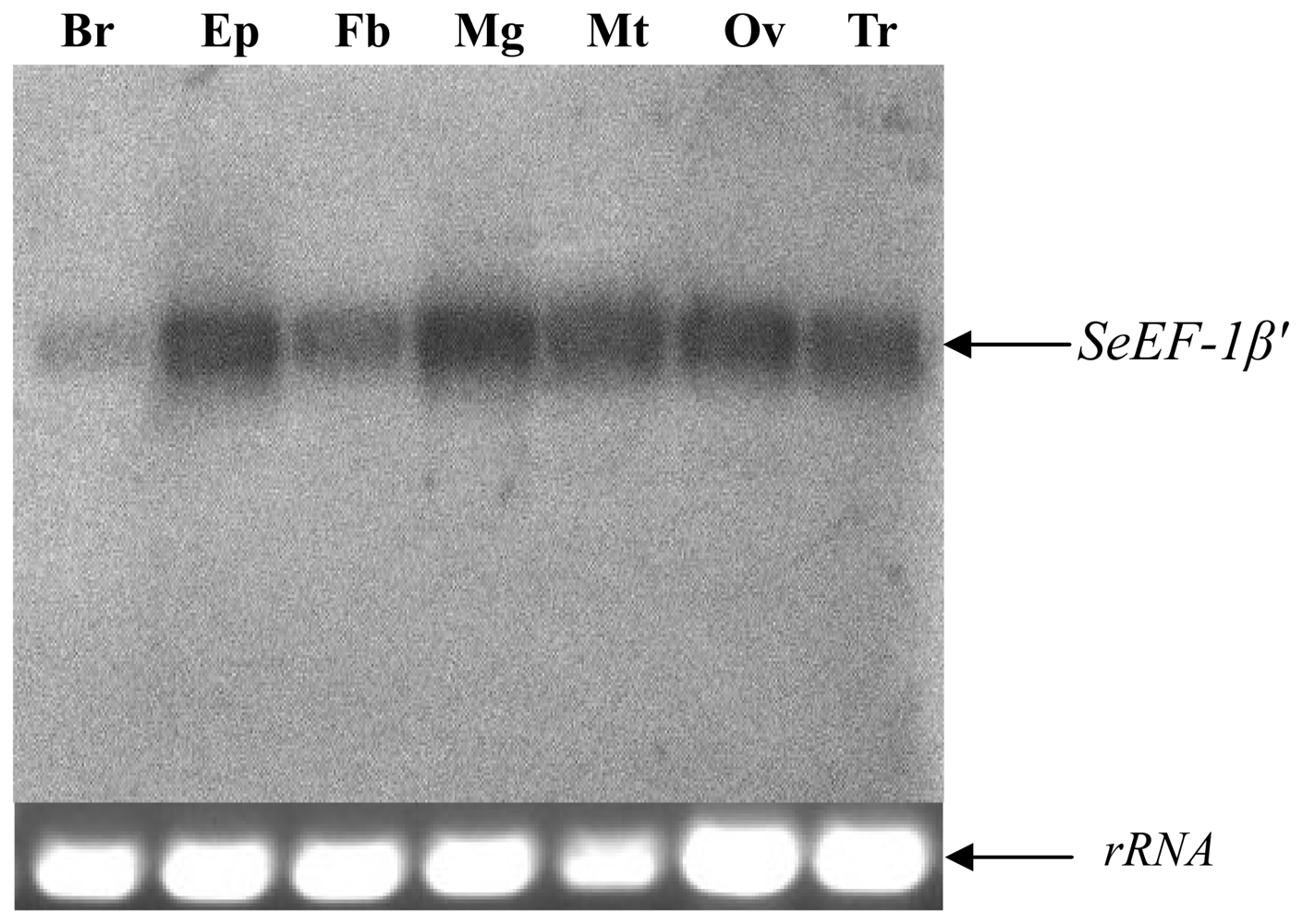
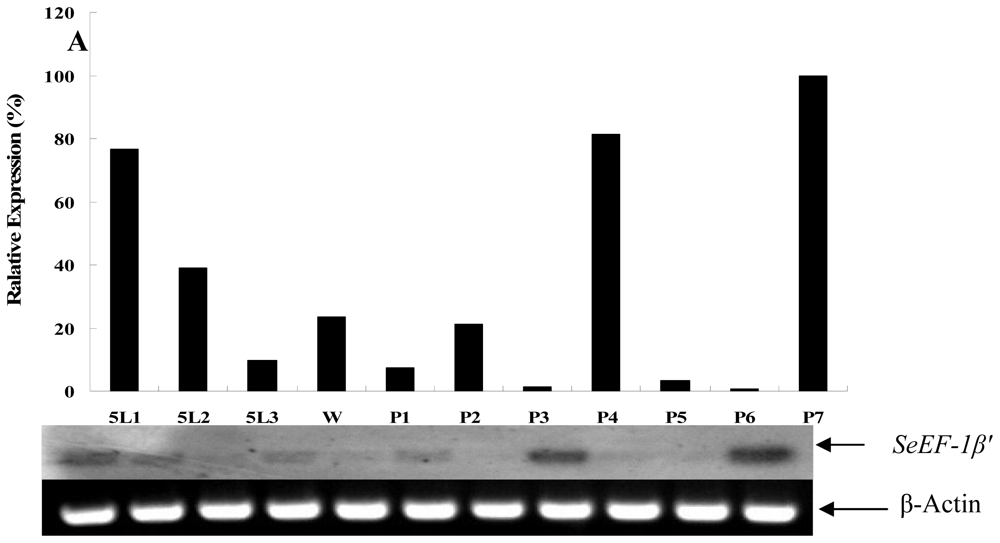
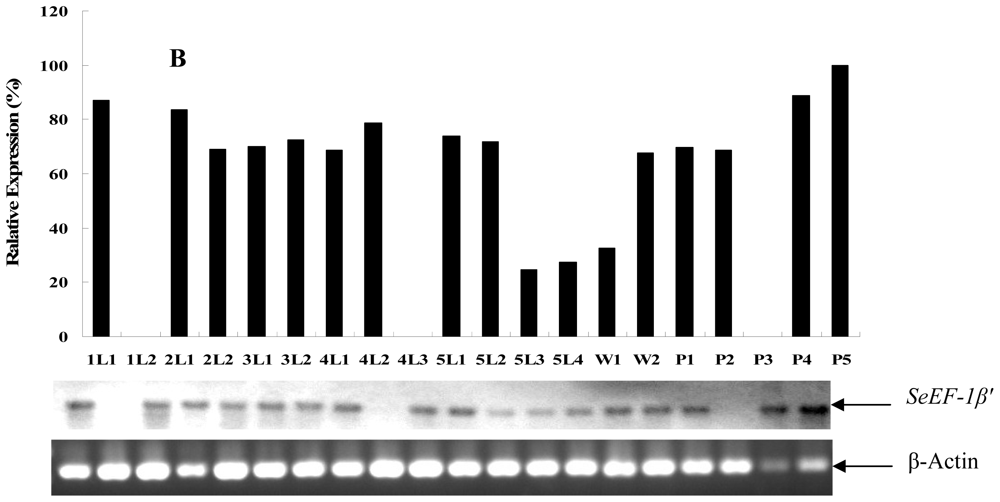
2.2. Tissue Distribution and Developmental Expression of SeEF-1β′
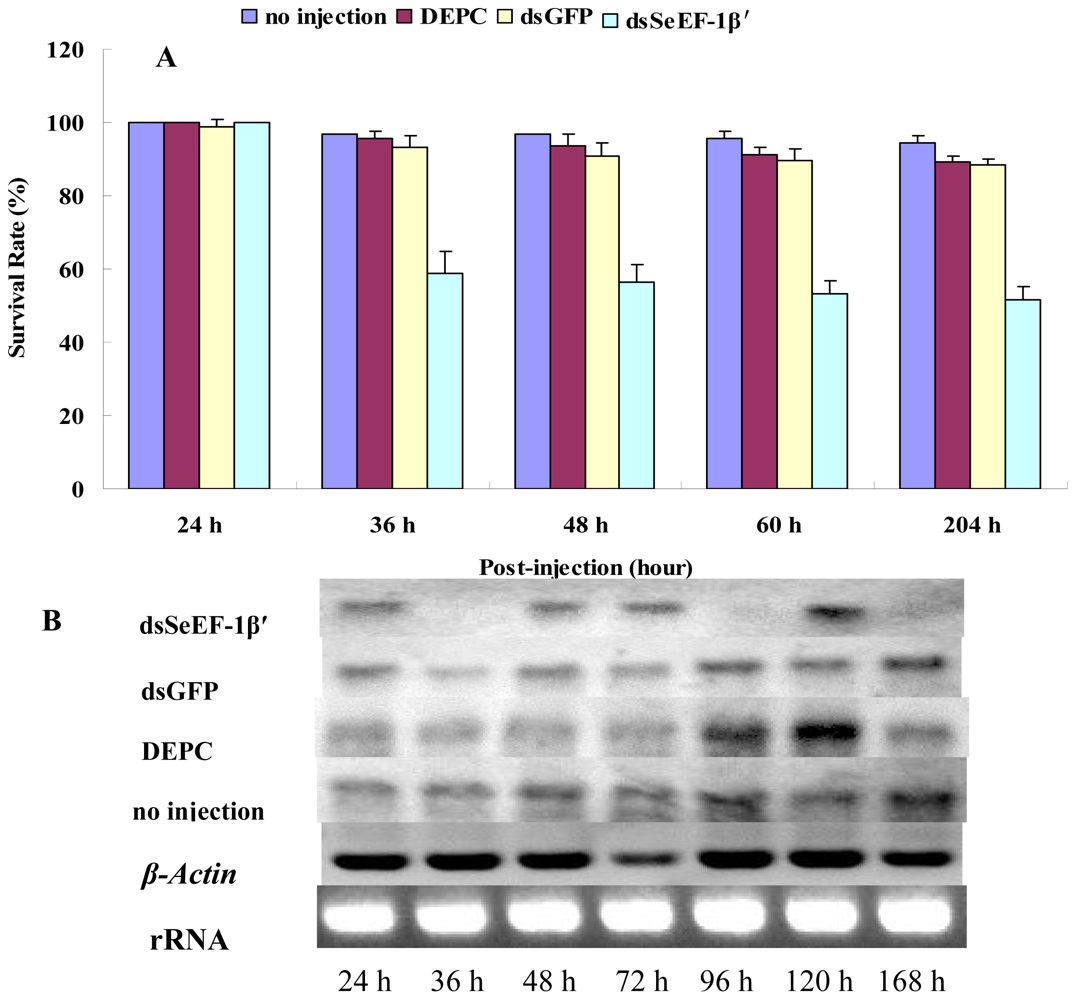
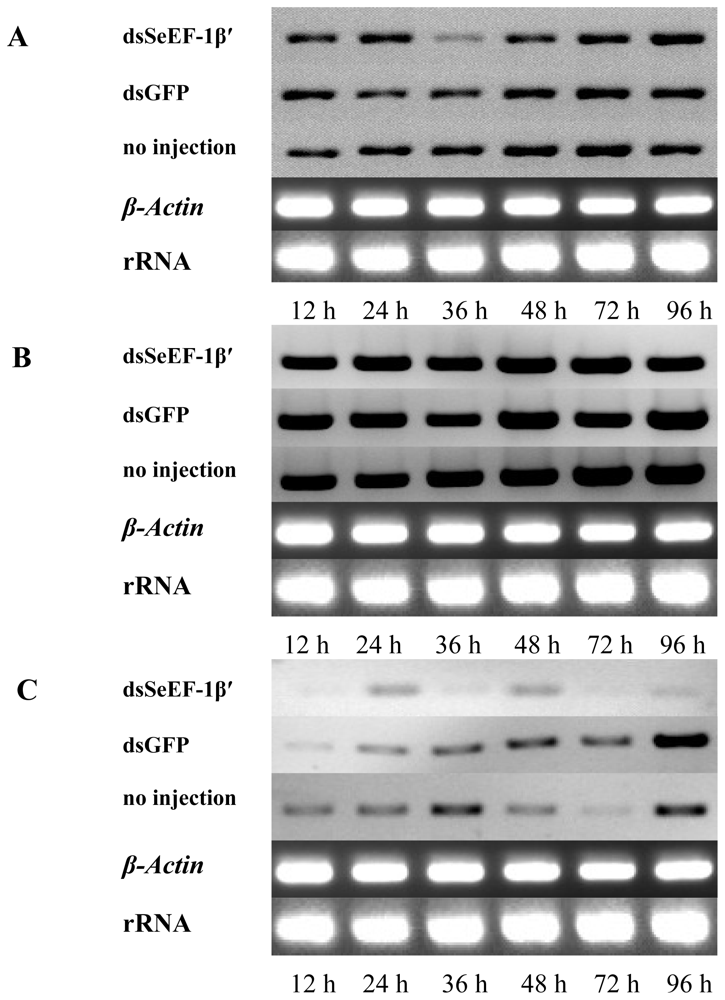
2.3. The Survival Rate and Expression of SeEF-1β′ after dsRNA Injection
2.4. The Expression of EF-1α, EF-2 and Fork Head mRNA after Injection of dsSeEF-1β′
3. Experimental Section
3.1. Insect Cultures
3.2. RNA Isolation, cDNA Synthesis and PCR
3.3. Rapid Amplification of cDNA Ends (RACE)
3.4. cDNA and Protein Sequence Analyses
3.5. Northern Blot
3.6. Determination of Developmental Expression of SeEF-1β′ by RT-PCR Analysis
3.7. Injection of dsSeEF-1β′ into S. exigua Larvae
3.8. Observation of Insect Survival and Data Analysis
3.9. RT-PCR Analysis of EF-1β′ Gene Silencing and EF-1α, EF-2 and Fork Head (FH) mRNA Expression
4. Conclusions
Acknowledgments
References
- Linz, J.E.; Sypherd, P.S. Expression of three genes for elongation factor 1α during morphogenesis of Mucor racemosus. Mol. Cell. Biol 1987, 7, 1925–1932. [Google Scholar]
- Riis, B.; Rattan, S.I.S.; Clark, B.F.C.; Merrick, W.C. Eukaryotic protein elongation factors. Trends Biochem. Sci 1990, 15, 420–424. [Google Scholar]
- Bassel, G.J.; Powers, C.M.; Taneja, K.L.; Singer, R.H. Single mRNAs visualized by ultrastructural in situ hybridization are principally localized at actin filament intersections in fibroblasts. J. Cell Biol 1994, 126, 863–876. [Google Scholar]
- Margutti, P.; Ortona, E.; Vaccari, S.; Barca, S.; Riganò, R.; Teggi, A.; Muhschlegel, F.; Frosch, M.; Siracusano, A. Cloning and expression of a cDNA encoding an elongation factor 1β/δ protein from Echinococcus granulosus with immunogenic activity. Parasite Immunol 1999, 21, 485–492. [Google Scholar]
- Fujita, T.; Piuz, I.; Schlegel, W. The transcription elongation factors NELF, DSIF and P-TEFb control constitutive transcription in a gene-specific manner. FEBS Lett 2009, 583, 2893–2898. [Google Scholar]
- Ibba, M.; Becker, H.D.; Stathopoulos, C.; Tumbula, D.L.; Söll, D. The adaptor hypothesis revisited. Trends Biochem. Sci 2000, 25, 311–316. [Google Scholar]
- Roy, H.; Becker, H.D.; Mazauric, M.H.; Kern, D. Structural elements defining elongation factor Tu mediated suppression of codon ambiguity. Nucleic Acids Res 2007, 35, 3420–3430. [Google Scholar]
- Krumm, A.; Meulia, T.; Groudine, M. Common mechanisms for the control of eukaryotic transcriptional elongation. Bioessays 1993, 15, 659–665. [Google Scholar]
- Uptain, S.M.; Kane, C.M.; Chamberlin, M.J. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem 1997, 66, 117–172. [Google Scholar]
- Orphanides, G.; Reinberg, D. A unified theory of gene expression. Cell 2002, 108, 439–451. [Google Scholar]
- Baugh, L.R.; Demodena, J.; Sternberg, P.W. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science 2009, 324, 92–94. [Google Scholar]
- Bourne, H.R.; Sanders, D.A.; McCormick, F. The GTPase superfamily: Conserved structure and molecular mechanism. Nature 1991, 349, 117–127. [Google Scholar]
- Dhandayuthapani, S.; Banu, M.J.; Kashiwabara, Y. Cloning and sequence determination of the gene coding for the elongation factor Tu of Mycobacterium lepra. J. Biochem 1994, 115, 664–669. [Google Scholar]
- Nechifor, R.; Murataliev, M.; Wilson, K.S. Functional interactions between the G′ subdomain of bacterial translation factor EF-G and ribosomal protein L7/L12. J. Biol. Chem 2007, 282, 36998–37005. [Google Scholar]
- Bunai, F.; Ando, K.; Ueno, H.; Numata, O. Tetrahymena eukaryotic translation elongation factor 1A (eEF1A) bundles filamentous actin through dimer formation. J. Biochem 2006, 140, 393–399. [Google Scholar]
- Bosch, L.B.; Kraal, J.M.; Van, N.J.; Van, D.A.; Talens, A.; Vijgenboom, E. Novel RNA interactions with elongation factor EF-Tu: consequences for protein synthesis and gene expression. Trends Biochem. Sci 1985, 10, 313–316. [Google Scholar]
- Browning, K.S. The plant translational apparatus. Plant Mol. Biol 1996, 32, 107–144. [Google Scholar]
- Kamiie, K.; Nomura, Y.; Kobayashi, S.; Taira, H.; Kobayashi, K.; Yamashita, T.; Kidou, S.; Ejiri, S. Cloning and expression of Bombyx mori silk gland elongation factor 1γ in Escherichia coli. Biosci. Biotechnol. Biochem 2002, 66, 558–565. [Google Scholar]
- Ejiri, S.; Saito, K.; Nakamura, H.; Kawasaki, H.; Katsumata, T. Proceedings of the International Symposium “Molecular Organization of Biological Structure”, Moscow, USSR, 24–27 June 1989; p. 238.
- Van Damme, H.T.; Amons, R.; Karssies, R.; Timmers, C.J.; Janssen, G.M.; Möller, W. Elongation factor 1β of artemia: Localization of functional sites and homology to elongation factor 1δ. Biochim. Biophys. Acta 1990, 1050, 241–247. [Google Scholar]
- Janssen, G.M.C.; Möller, W. Elongation factor 1βγ fromArtemia. Purification and properties of its subunits. Eur. J. Biochem 1988, 171, 119–129. [Google Scholar]
- Yang, F.; Demma, M.; Warren, V.; Dharmawardhane, S.; Condeelis, J. Identification of an actin-binding protein from Dictyostelium as elongation factor 1α. Nature 1990, 347, 494–496. [Google Scholar]
- Gross, S.R.; Kinzy, T.G. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat. Struct. Mol. Biol 2005, 12, 772–778. [Google Scholar]
- Kiel, J.A.; Titorenko, V.I.; van der Klei, I.J.; Veenhuis, M. Overproduction of translation elongation factor 1α (eEF1A) suppresses the peroxisome biogenesis defect in a Hansenula polymorpha pex3 mutant via translational read-through. FEMS Yeast Res 2007, 7, 1114–1125. [Google Scholar]
- Kim, S.; Kellner, J.; Lee, C.H.; Coulombe, P.A. Interaction between the keratin cytoskeleton and eEF1B γ affects protein synthesis in epithelial cells. Nat. Struct. Mol. Biol 2007, 14, 982–983. [Google Scholar]
- Valouev, I.A.; Fominov, G.V.; Sokolova, E.E.; Smirnov, V.N.; Ter-Avanesyan, M.D. Elongation factor eEF1B modulates functions of the release factors eRF1 and eRF3 and the efficiency of translation termination in yeast. BMC Mol. Biol 2009, 10. [Google Scholar] [CrossRef]
- Taira, H.; Kamiie, K.; Kakuta, A.; Ooura, H.; Matsumoto, S.; Ejiri, S.; Katsumata, T. Nucleotide sequence of the cDNA encoding silk gland elongation factor 1β′. Nucleic Acids Res 1992, 20, 6734. [Google Scholar]
- Janssen, G.M.C.; Möller, W. Elongation factor 1βγ from Artemia. Purification and properties of its subunits. Eur. J. Biochem 1988, 171, 119–129. [Google Scholar]
- Kamiie, K.; Taira, H.; Ooura, H.; Kakuta, A.; Matsumoto, S.; Ejiri, S.; Katsumata, T. Nucleotide sequence of the cDNA encoding silk gland elongation factor 1 alpha. Nucleic Acids Res 1993, 21, 742. [Google Scholar]
- Wang, X.; Lee, C.; Gilmour, D.S.; Gergen, J.P. Transcription elongation controls cell fate specification in the Drosophila embryo. Genes Dev 2007, 21, 1031–1036. [Google Scholar]
- Gao, L.; Zuo, H.; Liu, K.; Li, H.; Zhong, G. A new strategy for identification of highly conserved microRNAs in non-model insect, Spodoptera litura. Int. J. Mol. Sci 2012, 13, 612–627. [Google Scholar]
- Tang, B.; Chen, X.F.; Liu, Y.; Tian, H.G.; Liu, J.; Hu, J.; Xu, W.H.; Zhang, W.Q. Characterization and expression patterns of a membrane-bound trehalase from Spodoptera exigua. BMC Mol. Biol 2008, 9, 51. [Google Scholar]
- Tang, B.; Zheng, H.Z.; Xu, Q.; Zhou, Q.; Wang, G.J.; Zhang, F.; Wang, S.G.; Zhang, Z.H. Cloning and pattern of expression of trehalose-6-phosphate synthase cDNA from Catantops pinguis (Orthoptera: Catantopidae). Eur. J. Entomol 2011, 108, 355–363. [Google Scholar]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem 1987, 162, 156–159. [Google Scholar]
- Lv, L.-L.; Duan, J.; Xie, J.-H.; Liu, Y.-G.; Wei, C.-B.; Liu, S.-H.; Zhang, J.-X.; Sun, G.-M. Cloning and Expression Analysis of a PISTILLATA Homologous Gene from Pineapple (Ananas comosus L. Merr). Int. J. Mol. Sci 2012, 13, 1039–1053. [Google Scholar]
- Meng, X.; Xu, Z.; Song, R. Molecular cloning and characterization of a vacuolar H(+)-pyrophosphatase from Dunaliella viridis. Mol. Biol. Rep 2011, 38, 3375–3382. [Google Scholar]
- Sun, Y.; Lin, H.-D.; Tang, W.-Q.; Ju, Y.-M.; Liu, Z.-Z.; Liu, D.; Yang, J.-Q. Polymorphic microsatellite loci isolated from the Squalidus argentatus using PCR-based isolation of microsatellite arrays (PIMA). Int. J. Mol. Sci 2011, 12, 5666–5671. [Google Scholar]
- Swiss Institute of Bioinformatics. ExPASy Proteomics website. Available online: http://expasy.org/ accessed on 28 June 2012.
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucl. Acids Res. 1988, 16, pp. 10881–10890. Available online: http://multalin.toulouse.inra.fr/multalin/multalin.html accessed on 28 June 2012.
- Choo, Y.M.; Lee, K.S.; Kim, B.Y.; Kim, D.H.; Yoon, H.J.; Sohn, H.D.; Jin, B.R. A gut-specific chitinase from the mulberry longicorn beetle, Apriona germari (Coleoptera: Cerambycidae): cDNA cloning, gene structure, expression and enzymatic activity. Eur. J. Entomol 2007, 104, 173–180. [Google Scholar]
- Chen, X.F.; Tian, H.G.; Zou, L.Z.; Tang, B.; Hu, J.; Zhang, W.Q. Disruption of Spodoptera exigua larval development by silencing chitin synthase gene A with RNA interference. Bull. Entomol. Res 2008, 98, 613–619. [Google Scholar]
- Zhang, T.Y.; Sun, J.S.; Zhang, Q.R.; Xu, J.; Jiang, R.J.; Xu, W.H. The diapause hormone-pheromone biosynthesis activating neuropeptide gene of Helicoverpa armigera encodes multiple peptides that break, rather than induce, diapause. J. Insect Physiol 2004, 50, 547–554. [Google Scholar]



© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhao, L.-N.; Qin, Z.; Wei, P.; Guo, H.-S.; Dang, X.-L.; Wang, S.-G.; Tang, B. Elongation Factor 1β' Gene from Spodoptera exigua: Characterization and Function Identification through RNA Interference. Int. J. Mol. Sci. 2012, 13, 8126-8141. https://doi.org/10.3390/ijms13078126
Zhao L-N, Qin Z, Wei P, Guo H-S, Dang X-L, Wang S-G, Tang B. Elongation Factor 1β' Gene from Spodoptera exigua: Characterization and Function Identification through RNA Interference. International Journal of Molecular Sciences. 2012; 13(7):8126-8141. https://doi.org/10.3390/ijms13078126
Chicago/Turabian StyleZhao, Li-Na, Zi Qin, Ping Wei, Hong-Shuang Guo, Xiang-Li Dang, Shi-Gui Wang, and Bin Tang. 2012. "Elongation Factor 1β' Gene from Spodoptera exigua: Characterization and Function Identification through RNA Interference" International Journal of Molecular Sciences 13, no. 7: 8126-8141. https://doi.org/10.3390/ijms13078126
APA StyleZhao, L.-N., Qin, Z., Wei, P., Guo, H.-S., Dang, X.-L., Wang, S.-G., & Tang, B. (2012). Elongation Factor 1β' Gene from Spodoptera exigua: Characterization and Function Identification through RNA Interference. International Journal of Molecular Sciences, 13(7), 8126-8141. https://doi.org/10.3390/ijms13078126



