Identification of Late Embryogenesis Abundant (LEA) Protein Putative Interactors Using Phage Display
Abstract
:1. Introduction
2. Results and Discussion
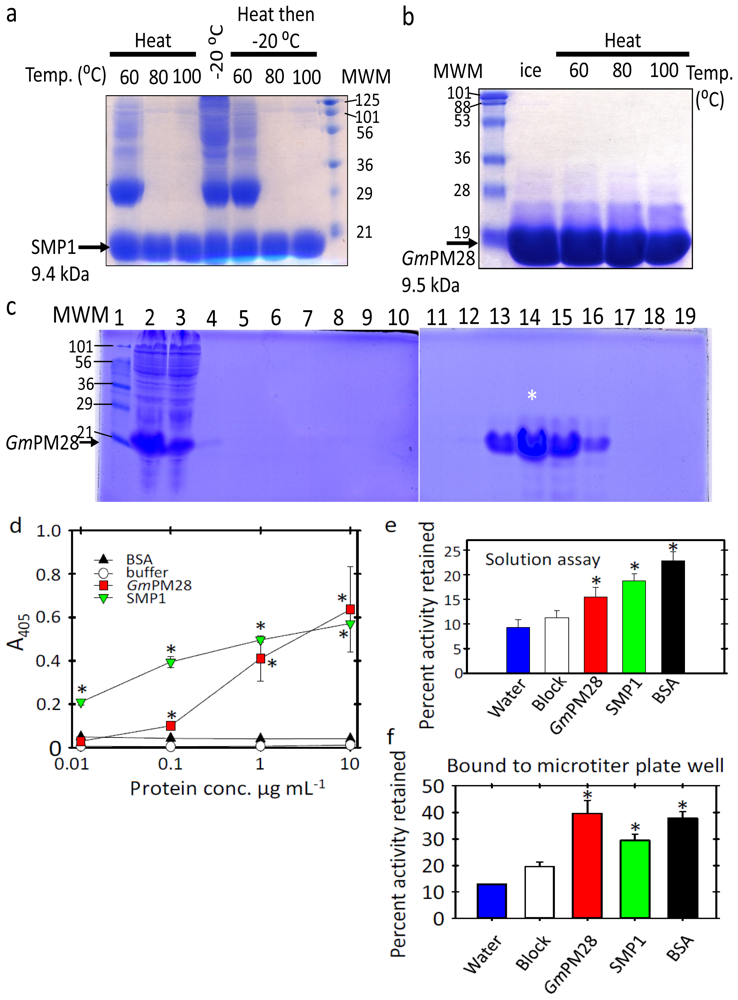
2.1. The Two LEA Proteins Are Boiling-Stable Proteins
2.2. Both LEA Proteins Afford Limited Protection to GLUCOSE-6-PHOSPHATE DEHYDROGENASE against Heat Stress
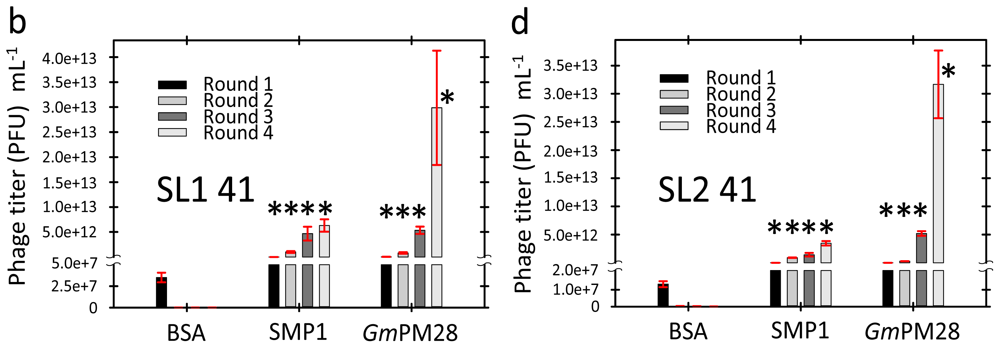
2.3. Phage Was Retained by Both LEA Proteins
2.4. LEA Protein-Retained Phage Contained Insert
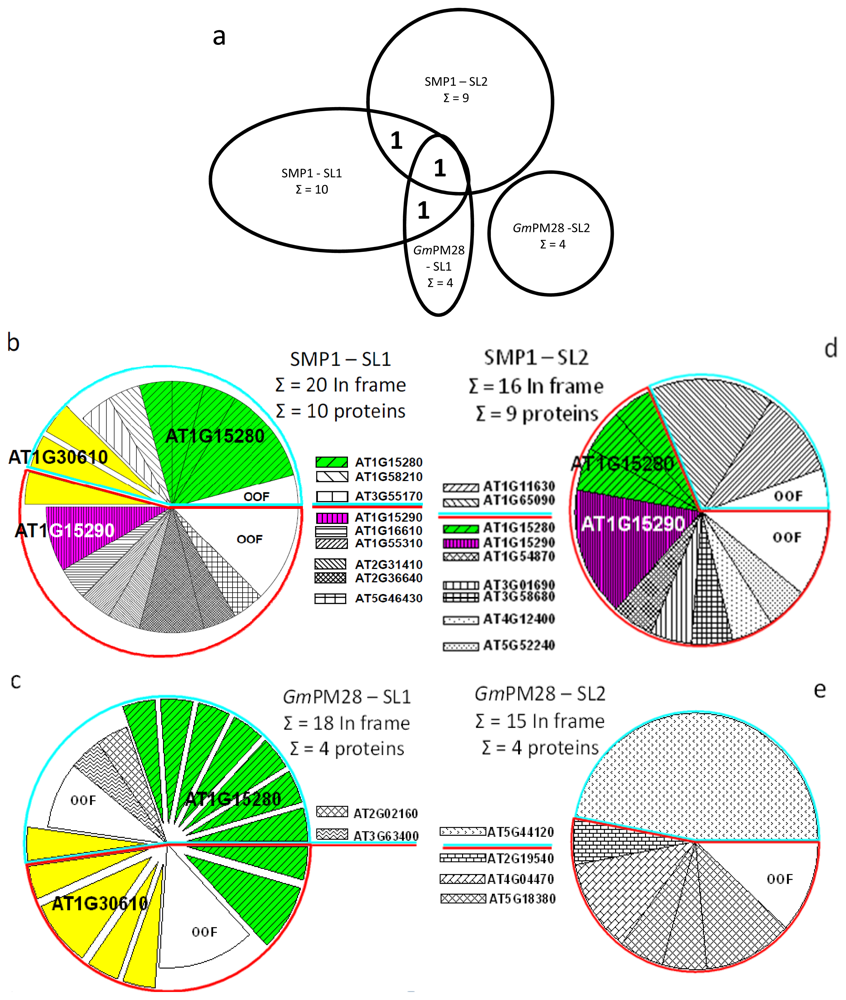
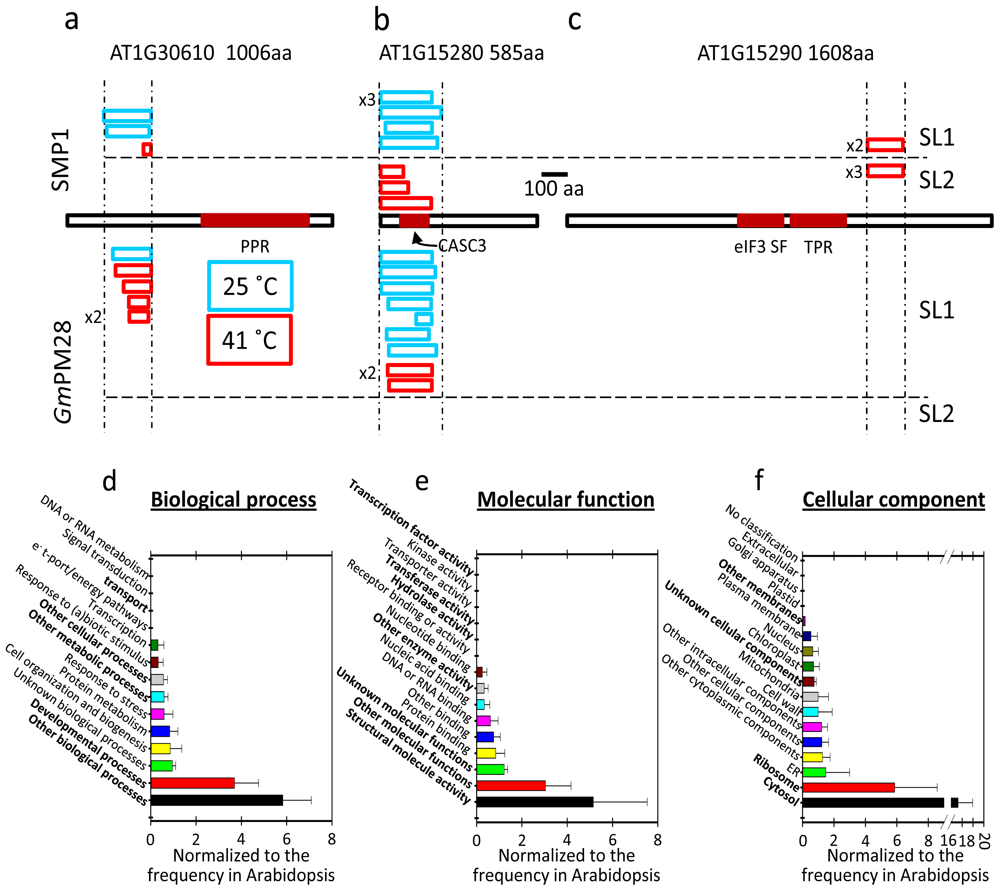
2.5. The Two LEA Protein Homologues Target Discrete Proteins, Some of Which Are Identical
2.6. Proteins Resident in the Cytoplasm and Associated with the Translational Machinery Are Over-Represented among the Retrieved Targets
3. Experimental Section
3.1. Cloning SMP1 and GmPM28
3.2. Recombinant Protein Production and Purification
3.3. Characterization of SMP1 and GmPM28 as Boiling-Stable Proteins
3.4. Determination of Recombinant SMP1 and GmPM28 Capacity to Bind to Microtiter Plate Wells
3.5. GLUCOSE 6 PHOSPHATE DEHYDROGENASE (G6PDH) Protection Assays
3.6. Biopanning
3.7. Titering
3.8. Plaque Isolation, PCR and Sequencing
3.9. Statistical Analysis
4. Conclusions
Supplemental Information
ijms-13-06582-s001.pdfAcknowledgments
References
- Dure, L., III; Greenway, S.C.; Galau, G.A. Developmental biochemistry of cottonseed embryogenesis and germination: Changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry 1981, 20, 4162–4168. [Google Scholar]
- Galau, G.A.; Hughes, D.W.; Dure, L.I. Developmental biochemistry of cottonseed embryogenesis and germination: Changing messenger ribonucleic acid populations as shown by reciprocal heterologous complementary deoxyribonucleic acid-messenger ribonucleic acid hybridizationembryogenesis-abundant (Lea) mRNAs. Plant Mol. Biol 1986, 7, 155–170. [Google Scholar]
- Garay-Arroyo, A.; Colmenero-Flores, J.M.; Garciarrubio, A.; Covarrubias, A.A. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J. Biol. Chem 2000, 275, 5668–5674. [Google Scholar]
- Wise, M.J. LEAping to conclusions: A computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinforma 2003, 4. [Google Scholar] [CrossRef] [Green Version]
- Mouillon, J.M.; Gustafsson, P.; Harryson, P. Structural investigation of disordered stress proteins. Comparison of full-length dehydrins with isolated peptides of their conserved segments. Plant Physiol 2006, 141, 638–650. [Google Scholar]
- Hand, S.C.; Menze, M.A.; Toner, M.; Boswell, L.; Moore, D. LEA proteins during water stress: Not just for plants anymore. Annu. Rev. Physiol 2011, 73, 115–134. [Google Scholar]
- Tunnacliffe, A.; Wise, M.J. The continuing conundrum of the LEA proteins. Naturwissenschaften 2007, 94, 791–812. [Google Scholar]
- Cheng, Z.; Targolli, J.; Huang, X.; Wu, R. Wheat LEA genes, PMA80 and PMA1959, enhance dehydration tolerance of transgenic rice (Oryza sativa L.). Mol. Breed 2002, 10, 71–82. [Google Scholar]
- Brini, F.; Hanin, M.; Lumbreras, V.; Amara, I.; Khoudi, H.; Hassairi, A.; Pages, M.; Masmoudi, K. Overexpression of wheat dehydrin DHN-5 enhances tolerance to salt and osmotic stress in Arabidopsis thaliana. Plant Cell Rep 2007, 26, 2017–2026. [Google Scholar]
- Xiao, B.; Huang, Y.; Tang, N.; Xiong, L. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet 2007, 115, 35–46. [Google Scholar]
- Xu, D.; Duan, X.; Wang, B.; Hong, B.; Ho, T.; Wu, R. Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol 1996, 110, 249–257. [Google Scholar]
- Manfre, A.J.; Lanni, L.M.; Marcotte, W.R., Jr. The Arabidopsis group 1 LATE EMBRYOGENESIS ABUNDANT protein ATEM6 is required for normal seed development. Plant Physiol. 2006, 140, 140–149. [Google Scholar]
- Saavedra, L.; Svensson, J.; Carballo, V.; Izmendi, D.; Welin, B.; Vidal, S. A dehydrin gene in Physcomitrella patens is required for salt and osmotic stress tolerance. Plant J 2006, 45, 237–249. [Google Scholar]
- Salleh, F.M.; Evans, K.; Goodall, B.; Machin, H.; Mowla, S.B.; Mur, L.A.; Runions, J.; Theodoulou, F.L.; Foyer, C.H.; Rogers, H.J. A novel function for a redox-related LEA protein (SAG21/AtLEA5) in root development and biotic stress responses. Plant Cell Environ 2012, 35, 418–429. [Google Scholar]
- Olvera-Carrillo, Y.; Campos, F.; Reyes, J.L.; Garciarrubio, A.; Covarrubias, A.A. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol 2010, 154, 373–390. [Google Scholar]
- Tompa, P.; Kovacs, D. Intrinsically disordered chaperones in plants and animals. Biochem. Cell Biol 2010, 88, 167–174. [Google Scholar]
- Kovacs, D.; Rakacs, M.; Agoston, B.; Lenkey, K.; Semrad, K.; Schroeder, R.; Tompa, P. Janus chaperones: Assistance of both RNA- and protein-folding by ribosomal proteins. FEBS Lett 2009, 583, 88–92. [Google Scholar]
- Goyal, K.; Walton, L.J.; Tunnacliffe, A. LEA proteins prevent protein aggregation due to water stress. Biochem. J 2005, 388, 151–157. [Google Scholar]
- Wise, M.J.; Tunnacliffe, A. POPP the question: What do LEA proteins do? Trends Plant Sci 2004, 9, 13–17. [Google Scholar]
- Reyes, J.L.; Rodrigo, M.-J.; Colmenero-Flores, J.M.; Gil, J.-V.; Garay-Arroyo, A.; Campos, F.; Salamini, F.; Bartels, D.; Covarrubias, A.A. Hydrophilins from distant organisms can protect enzymatic activities from water limitation effects in vitro. Plant Cell Environ 2005, 28, 709–718. [Google Scholar]
- Reyes, J.L.; Campos, F.; Wei, H.; Arora, R.; Yang, Y.; Karlson, D.T.; Covarrubias, A.A. Functional dissection of hydrophilins during in vitro freeze protection. Plant Cell Environ 2008, 31, 1781–1790. [Google Scholar]
- Olvera-Carrillo, Y.; Luis Reyes, J.; Covarrubias, A.A. Late embryogenesis abundant proteins: Versatile players in the plant adaptation to water limiting environments. Plant Signal Behav 2011, 6, 586–589. [Google Scholar]
- Hara, M.; Terashima, S.; Kuboi, T. Characterization and cryoprotective activity of cold-responsive dehydrin from Citrus unshiu. J. Plant Physiol 2001, 158, 1333–1339. [Google Scholar]
- Tolleter, D.; Hincha, D.K.; Macherel, D. A mitochondrial late embryogenesis abundant protein stabilizes model membranes in the dry state. Biochim. Biophys Acta 2010, 1798, 1926–1933. [Google Scholar] [Green Version]
- Shih, M.-D.; Hsieh, T.-Y.; Lin, T.-P.; Hsing, Y.C.; Hoekstra, F.A. Characterization of two soybean (Glycine max L.) LEA IV proteins by circular dichroism and fourier transform infrared spectrometry. Plant Cell Physiol 2010, 51, 395–407. [Google Scholar]
- Bies-Etheve, N.; Gaubier-Comella, P.; Debures, A.; Lasserre, E.; Jobet, E.; Raynal, M.; Cooke, R.; Delseny, M. Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol. Biol 2008, 67, 107–124. [Google Scholar]
- Hundertmark, M.; Hincha, D.K. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. In BMC Genomics; 2008; Volume 9. [Google Scholar] [CrossRef]
- Chen, T.; Nayak, N.; Majee, S.M.; Lowenson, J.; Schafermeyer, K.R.; Eliopoulos, A.C.; Lloyd, T.D.; Dinkins, R.; Perry, S.E.; Forsthoefel, N.R.; et al. Substrates of the Arabidopsis thaliana protein isoaspartyl methyltransferase1 identified using phage display and biopanning. J. Biol. Chem 2010, 285, 37281–37292. [Google Scholar]
- Hilhorst, H.W.M. Dose-response analysis of factors involved in germination and secondary dormancy of seeds of sisymbrium-officinale. 1. Phytochrome. Plant Physiol 1990a, 94, 1090–1095. [Google Scholar]
- Hilhorst, H.W.M. Dose-response analysis of factors involved in germination and secondary dormancy of seeds of sisymbrium-officinale. 2. Nitrate. Plant Physiol 1990b, 94, 1096–1102. [Google Scholar]
- Gubler, F.; Millar, A.A.; Jacobsen, J.V. Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol 2005, 8, 183–187. [Google Scholar]
- Hilhorst, H.W.M.; Toorop, P.E. Review on dormancy, germinability, and germination in crop and weed seeds. Adv. Agron 1997, 61, 111–165. [Google Scholar]
- Jung, S.; Honegger, A.; Pluckthun, A. Selection for improved protein stability by phage display. J. Mol. Biol 1999, 294, 163–180. [Google Scholar]
- Ritchie, D.A.; Malcolm, F.E. Heat-stable and density mutants of phages T1, T3 and T7. J. Gen. Virol 1970, 9, 35–43. [Google Scholar]
- Close, T.J.; Kortt, A.A.; Chandler, P.M. A cdna-based comparison of dehydration-induced proteins (dehydrins) in barley and corn. Plant Mol. Biol 1989, 13, 95–108. [Google Scholar]
- Kikawada, T.; Nakahara, Y.; Kanamori, Y.; Iwata, K.I.; Watanabe, M.; Mcgee, B.; Tunnacliffe, A.; Okuda, T. Dehydration-induced expression of LEA proteins in an anhydrobiotic chironomid. Biochem. Biophys. Res. Commun 2006, 348, 56–61. [Google Scholar]
- Li, C.; Clarke, S. A protein methyltransferase specific for altered aspartyl residues is important in escherichia-coli stationary-phase survival and heat-shock resistance. Proc. Natl. Acad. Sci. USA 1992, 89, 9885–9889. [Google Scholar]
- Clarke, S. Aging as war between chemical and biochemical processes: Protein methylation and the recognition of age-damaged proteins for repair. Ageing Res. Rev 2003, 2, 263–285. [Google Scholar]
- Lam, J.M.; Pwee, K.H.; Sun, W.Q.; Chua, Y.L.; Wang, X.J. Enzyme-stabilizing activity of seed trypsin inhibitors during desiccation. Plant Sci 1999, 142, 209–218. [Google Scholar]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Heazlewood, J.L.; Verboom, R.E.; Tonti-Filippini, J.; Small, I.; Millar, A.H. SUBA: The arabidopsis subcellular database. Nucleic Acids Res 2007, 35, D213–D218. [Google Scholar]
- van Bentem, S.D.; Anrather, D.; Dohnal, I.; Roitinger, E.; Csaszar, E.; Joore, J.; Buijnink, J.; Carreri, A.; Forzani, C.; Lorkovic, Z.J.; et al. Site-specific phosphorylation profiling of Arabidopsis proteins by mass spectrometry and peptide chip analysis. J. Proteome Res 2008, 7, 2458–2470. [Google Scholar]
- Yeung, E.C.; Meinke, D.W. Embryogenesis in angiosperms-development of the suspensor. Plant Cell 1993, 5, 1371–1381. [Google Scholar]
- Cushing, D.A.; Forsthoefel, N.R.; Gestaut, D.R.; Vernon, D.M. Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta 2005, 221, 424–436. [Google Scholar]
- Ascencio-Ibanez, J.T.; Sozzani, R.; Lee, T.J.; Chu, T.M.; Wolfinger, R.D.; Cella, R.; Hanley-Bowdoin, L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol 2008, 148, 436–454. [Google Scholar]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem 1951, 193, 265–275. [Google Scholar]
- Noltmann, E.A.; Gubler, C.J.; Kuby, S.A. Glucose 6-phosphate dehydrogenase (Zwischenferment). I. Isolation of the crystalline enzyme from yeast. J. Biol. Chem 1961, 236, 1225–1230. [Google Scholar]
- Kuramochi, K.; Miyano, Y.; Enomoto, Y.; Takeuchi, R.; Ishi, K.; Takakusagi, Y.; Saitoh, T.; Fukudome, K.; Manita, D.; Takeda, Y.; et al. Identification of small molecule binding molecules by affinity purification using a specific ligand immobilized on PEGA resin. Bioconjug. Chem 2008, 19, 2417–2426. [Google Scholar]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploetz, L.; et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res 2008, 36, D1009–D1014. [Google Scholar]

| In-Frame Hits of SMP1 at 25 °C Using SL1 after 4 Rounds of Panning | ||||
|---|---|---|---|---|
| Hits | Locus Identifier | Protein Identity | Independent Clones | Subcellular Residence/Authority |
| 6 | AT1G15280 | CASC3/Barentsz eIF4AIII binding | 4 | Predictors-SubLoc: nucleus; WoLFPSORT: nucleus |
| 2 | AT1G30610 | EMB88, EMB2279 pentatricopeptide (PPR) repeat-containing protein | 2 | Predictors-iPSORT: plastid; MitoPred: mitochondrion; MultiLoc: plastid; PeroxP: peroxisome; Predotar: plastid; SubLoc: nucleus; TargetP: plastid; WoLFPSORT: nucleus |
| 1 | AT1G58210 | EMB1674 | kinase interacting family protein | 1 | Predictors-iPSORT: mitochondrion; MitoPred: mitochondrion; MultiLoc: plastid; Predotar: mitochondrion; SubLoc: nucleus; TargetP: plastid; WoLFPSORT: nucleus |
| 1 | AT3G55170 | Ribosomal L29 family protein | 1 | SUBA MS/MS: cytosol: 17934214; plasma membrane: 19334764 Predictors: LOCtree: plastid; MitoPred: mitochondrion; SubLoc: nucleus; WoLFPSORT: nucleus |
| In-Frame Hits of GmPM28 at 25 °C Using SL1 after 4 Rounds of Panning | ||||
| Hits | Locus Identifier | Protein Identity | Independent Clones | Subcellular Residence/Authority |
| 7 | AT1G15280 | CASC3/Barentsz eIF4AIII binding | 7 | Predictors-SubLoc: nucleus; WoLFPSORT: nucleus |
| 1 | AT3G63400 | Cyclophilin-like peptidyl-prolyl cis-trans isomerasefamily protein | 1 | Predictors-SubLoc: nucleus |
| 1 | AT1G30610 | EMB88, EMB2279 | pentatricopeptide (PPR) repeat-containing protein | 1 | Predictors-iPSORT: plastid; MitoPred: mitochondrion; MultiLoc: plastid; PeroxP: peroxisome; Predotar: plastid; SubLoc: nucleus; TargetP: plastid; WoLFPSORT: nucleus |
| 1 | AT2G02160 | CCCH-type zinc finger family protein | 1 | Predictors-MitoPred: mitochondrion; SubLoc: nucleus; WoLFPSORT: nucleus |
| In-Frame Hits of SMP1 at 41 °C Using SL1 after 4 Rounds of Panning | ||||
| Hits | Locus Identifier | Protein Identity | Independent Clones | Subcellular Residence/Authority |
| 1 | AT1G30610 | EMB88, EMB2279 pentatricopeptide (PPR) repeat-containing protein | 1 | Predictors-iPSORT: plastid; MitoPred: mitochondrion; MultiLoc: plastid; PeroxP: peroxisome; Predotar: plastid; SubLoc: nucleus; TargetP: plastid; WoLFPSORT: nucleus |
| 3 | AT2G36640 | ATECP63, ECP63 embryonic cell protein 63 | 2 | Predictors-LOCtree: cytosol SubLoc: nucleus WoLFPSORT: peroxisome |
| 1 | AT1G55310 | SR33, SCL33, At-SCL33 | SC35-like splicing factor | 1 | Predictors-interchromatin granule, nuclear speck, nucleolus, plasma membrane/TAIR |
| 1 | AT5G46430 | Ribosomal protein L32e | 1 | SUBA MS/MS: cytosol: 17934214; cytosol: 21166475; plasma membrane: 19334764; plasma membrane: 15574830; Predictors-iPSORT: mitochondrion; LOCtree: mitochondrion; MitoPred: mitochondrion; Mitoprot 2: mitochondrion; MultiLoc: plastid; Predotar: mitochondrion; SubLoc: mitochondrion; TargetP: mitochondrion; WoLFPSORT: cytosol |
| 2 | AT1G15290 | Tetratricopeptide repeat (TPR)-like superfamily | 1 | SUBA MS/MS: cytosol: 21166475; Predictors-SubLoc: nucleus; WoLFPSORT: nucleus |
| 1 | AT2G31410 | unknown protein | 1 | Predictors-MitoPred: mitochondrion; SubLoc: nucleus; WoLFPSORT: nucleus |
| 1 | AT1G16610 | SR45, RNPS1 | arginine/serine-rich 45 | 1 | Mitochondria, nucleus plastid/BAR Cell eFP Browser |
| In-Frame Hits of GmPM28 at 41 °C Using SL1 after 4 Rounds of Panning | ||||
| Hits | Locus Identifier | Protein Identity | Independent Clones | Subcellular Residence/Authority |
| 5 | AT1G30610.1 | EMB88, EMB2279 | pentatricopeptide (PPR)repeatcontaining protein | 4 | Predictors-iPSORT: plastid; MitoPred: mitochondrion; MultiLoc: plastid; PeroxP: peroxisome; Predotar: plastid; SubLoc: nucleus; TargetP: plastid; WoLFPSORT: nucleus |
| 3 | AT1G15280.1 | CASC3/Barentsz eIF4AIII binding | 2 | Predictors-SubLoc: nucleus; WoLFPSORT: nucleus |
| In-Frame Hits of SMP1 at 25 °C Using SL2 after 4 Rounds of Panning | ||||
| Hits | Locus Identifier | Protein Identity | Independent Clones | Subcellular Residence/Authority |
| 2 | AT1G11630 | Pentatricopeptide repeat-containing protein | 2 | SUBA MS/MS: mitochondrion: 14671022; Predictors-iPSORT: mitochondrion; LOCtree: mitochondrion; MitoPred: mitochondrion; MultiLoc: plastid; Predotar : mitochondrion; SubLoc : nucleus; TargetP : mitochondrion; WoLFPSORT : plastid |
| 3 | AT1G65090 | unknown protein | 3 | cytosol, mitochondria/BAR Cell eFP Browser |
| In-Frame Hits of GmPM28 at 25 °C Using SL2 after 4 Rounds of Panning | ||||
| Hits | Locus Identifier | Protein Identity | Independent Clones | Subcellular Residence/Authority |
| 8 | AT5G44120 | CRA1, ATCRA1, CRU1 | RmlC-like cupins superfamily | 1 | SUBA MS/MS: plasma membrane; Predictors-Predotar: endoplasmic reticulum; SubLoc: nucleus; TargetP: extracellular; WoLFPSORT: vacuole |
| In-Frame Hits of SMP1 at 41 °C Using SL2 after 4 Rounds of Panning | ||||
| Hits | Locus Identifier | Protein Identity | Independent Clones | Subcellular Residence/Authority |
| 3 | AT1G15280.2 | CASC3/Barentsz eIF4AIII binding | 3 | Predictors-SubLoc: nucleus; WoLFPSORT: nucleus |
| 3 | AT1G15290.1 | Tetratricopeptide repeat (TPR)-like superfamily | 1 | SUBA MS/MS: cytosol: 21166475; Predictors-SubLoc: nucleus; WoLFPSORT: nucleus |
| 1 | AT4G12400.2 | Hop3|stress-inducible protein, putative | 1 | No information |
| 1 | AT1G54870.1 | NAD(P)-binding Rossmann-fold superfamily protein | 1 | Predictors-MultiLoc: mitochondrion; SubLoc: cytosol; WoLFPSORT: plastid |
| 1 | AT3G58680.1 | MBF1B, ATMBF1B | multiprotein bridging factor 1B | 1 | GFP: cytosol: 15610358; nucleus: 15610358; Annotators-AmiGO: nucleus; Predictors-LOCtree: cytosol; MitoPred: mitochondrion; Predotar: plastid; SubLoc: nucleus; WoLFPSORT: nucleus |
| 1 | AT3G01690.1 | alpha/beta-Hydrolases superfamily protein | 1 | Predictors-iPSORT: mitochondrion; SubLoc: nucleus; TargetP: plastid; WoLFPSORT: cytosol |
| 1 | AT5G52240.1 | MSBP1, ATMP1, AtMAPR5 membrane steroid binding protein1 | 1 | SUBA MS/MS: endoplasmic reticulum: 16618929; plasma membrane: 17317660; plasma membrane: 19334764; plasma membrane: 17644812; Annotators-AmiGO: plasma membrane; Predictors-SubLoc: cytosol; TargetP: extracellular; WoLFPSORT: endoplasmic reticulum |
| In-Frame Hits of GmPM28 at 41 °C Using SL2 after 4 Rounds of Panning | ||||
| Hits | Locus Identifier | Protein Identity | Independent Clones | Subcellular Residence/Authority |
| 4 | AT5G18380 | Ribosomal protein S5 domain 2-like superfamily | 3 | Annotators-UniProt : cytosol Predictors-iPSORT: plastid; LOCtree: cytosol; Mitoprot 2: mitochondrion; SubLoc: mitochondrion; WoLFPSORT: cytosol |
| 2 | AT4G04470 | PMP22| Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein | 1 | SUBA MS/MS: peroxisome: 18931141; plasma membrane: 19334764; Annotators-TAIR : peroxisome; AmiGO: peroxisome Predictors-iPSORT: mitochondrion; LOCtree: mitochondrion; MitoPred: mitochondrion; Mitoprot 2: mitochondrion; MultiLoc: mitochondrion; PeroxP: no data; Predotar: no data; SubLoc: mitochondrion; TargetP: mitochondrion; WoLFPSORT: plastid |
| 1 | AT2G19540 | Transducin family protein/WD-40 repeat family protein | 1 | Predictors-SubLoc: cytosol; WoLFPSORT: mitochondrion |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kushwaha, R.; Lloyd, T.D.; Schäfermeyer, K.R.; Kumar, S.; Downie, A.B. Identification of Late Embryogenesis Abundant (LEA) Protein Putative Interactors Using Phage Display. Int. J. Mol. Sci. 2012, 13, 6582-6603. https://doi.org/10.3390/ijms13066582
Kushwaha R, Lloyd TD, Schäfermeyer KR, Kumar S, Downie AB. Identification of Late Embryogenesis Abundant (LEA) Protein Putative Interactors Using Phage Display. International Journal of Molecular Sciences. 2012; 13(6):6582-6603. https://doi.org/10.3390/ijms13066582
Chicago/Turabian StyleKushwaha, Rekha, Taylor D. Lloyd, Kim R. Schäfermeyer, Santosh Kumar, and Allan Bruce Downie. 2012. "Identification of Late Embryogenesis Abundant (LEA) Protein Putative Interactors Using Phage Display" International Journal of Molecular Sciences 13, no. 6: 6582-6603. https://doi.org/10.3390/ijms13066582




