Structural Features of Caspase-Activating Complexes
Abstract
:1. Introduction
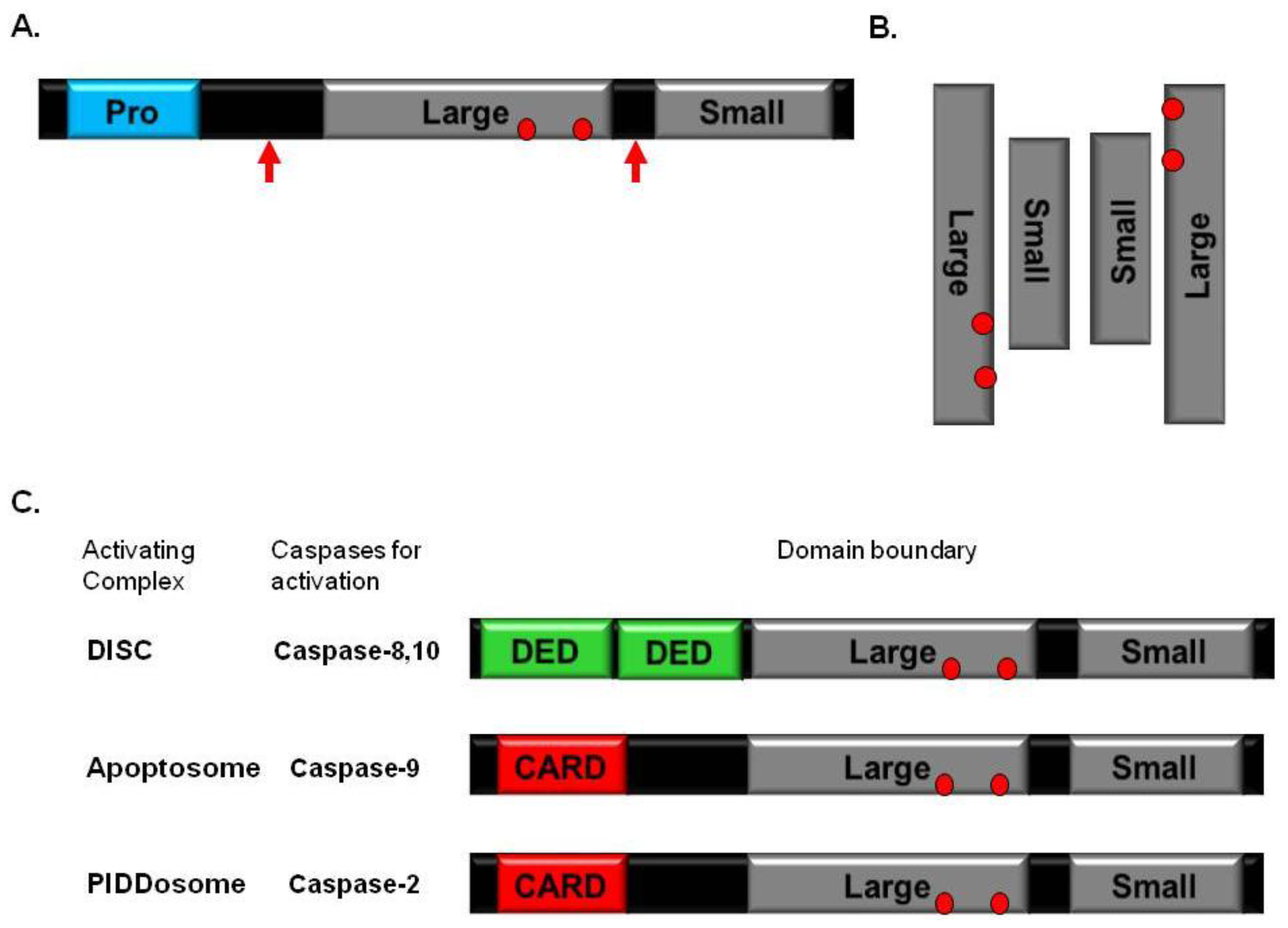
2. Caspase
3. Caspase-Activating Complexes and Death Domain Superfamily
4. DISC
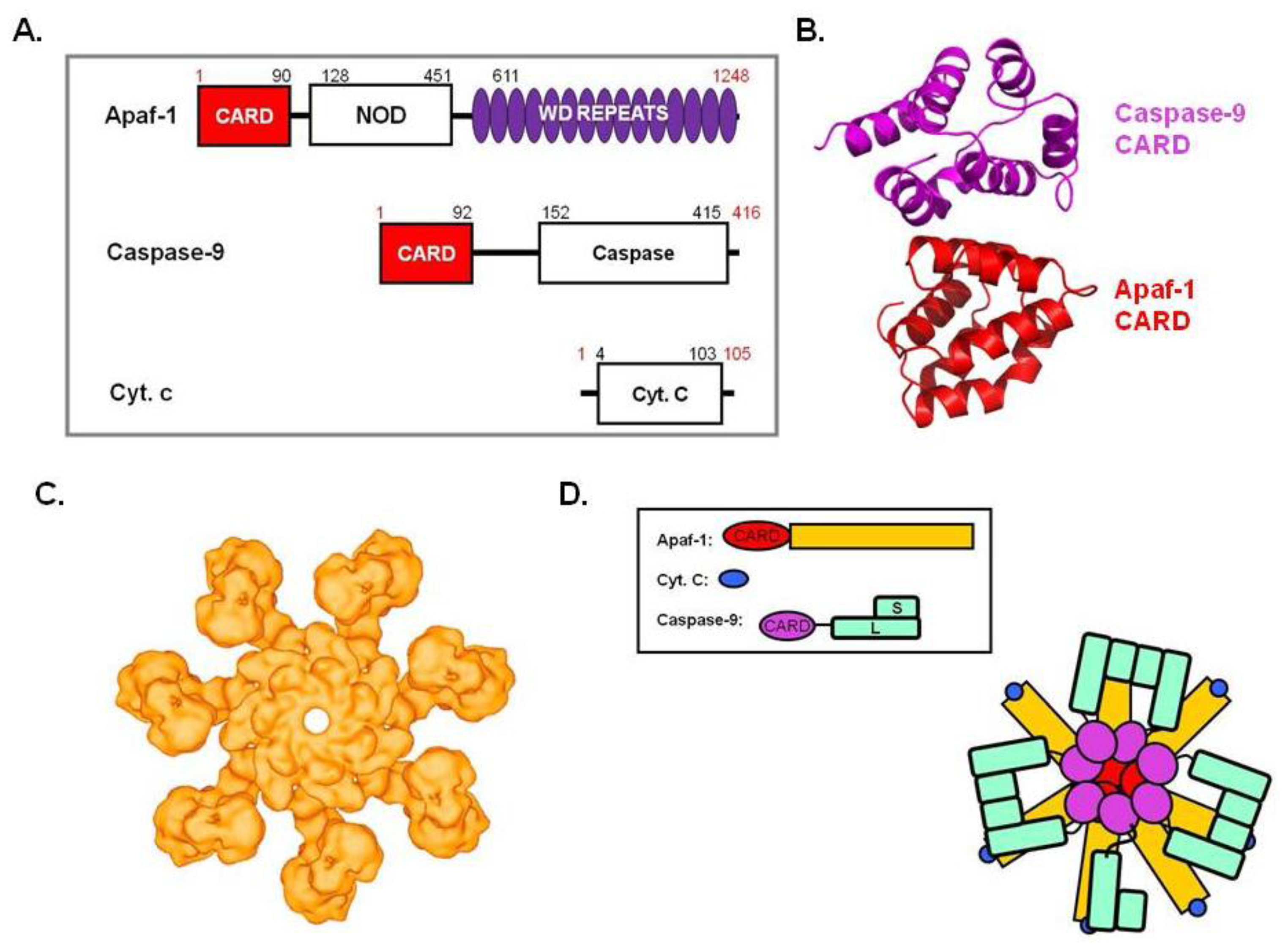
5. Apoptosome
6. PIDDosome
7. Conclusions
Acknowledgments
References
- Yeh, W.C.; Pompa, J.L.; McCurrach, M.E.; Shu, H.B.; Elia, A.J.; Shahinian, A.; Ng, M.; Wakeham, A.; Khoo, W.; Mitchell, K.; et al. FADD: Essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science 1998, 279, 1954–1958. [Google Scholar]
- Rathmell, J.C.; Thompson, C.B. Pathways of apoptosis in lymphocyte development, homeostasis, and disease. Cell 2002, 109, S97–S107. [Google Scholar]
- Strasser, A.; Jost, P.J.; Nagata, S. The many roles of FAS receptor signaling in the immune system. Immunity 2009, 30, 180–192. [Google Scholar]
- Kerr, J.F.; Wyllie, A.H.; Currie, A.R. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar]
- Duvall, E.; Wyllie, A.H. Death and the cell. Immunol. Today 1986, 7, 115–119. [Google Scholar]
- Rieux-Laucat, F.; Fischer, A.; Deist, F.L. Cell-death signaling and human disease. Curr. Opin. Immunol 2003, 15, 325–331. [Google Scholar]
- Oliveira, J.B.; Gupta, S. Disorders of apoptosis: Mechanisms for autoimmunity in primary immunodeficiency diseases. J. Clin. Immunol 2008, 28, S20–S28. [Google Scholar]
- Pawlowski, K.; Pio, F.; Chu, Z.; Reed, J.C.; Godzik, A. PAAD—A new protein domain associated with apoptosis, cancer and autoimmune diseases. Trends Biochem. Sci 2001, 26, 85–87. [Google Scholar]
- Ambrosini, G.; Adida, C.; Altieri, D.C. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med 1997, 3, 917–921. [Google Scholar]
- Andersen, M.H.; Becker, J.C.; Straten, P. Regulators of apoptosis: suitable targets for immune therapy of cancer. Nat. Rev. Drug Discov 2005, 4, 399–409. [Google Scholar]
- Evan, G.I.; Vousden, K.H. Proliferation, cell cycle and apoptosis in cancer. Nature 2001, 411, 342–348. [Google Scholar]
- Logue, S.E.; Gustafsson, A.B.; Samali, A.; Gottlieb, R.A. Ischemia/reperfusion injury at the intersection with cell death. J. Mol. Cell. Cardiol 2005, 38, 21–33. [Google Scholar]
- Salvesen, G.S.; Dixit, V.M. Caspases: Intracellular signaling by proteolysis. Cell 1997, 91, 443–446. [Google Scholar]
- Yang, X.; Chang, H.Y.; Baltimore, D. Autoproteolytic activation of pro-caspases by oligomerization. Mol. Cell 1998, 1, 319–325. [Google Scholar]
- Stennicke, H.R.; Salvesen, G.S. Catalytic properties of the caspases. Cell Death Differ 1999, 6, 1054–1059. [Google Scholar]
- Donepudi, M.; Grutter, M.G. Structure and zymogen activation of caspases. Biophys. Chem 2002, 101–102, 145–153. [Google Scholar]
- Park, H.H. Structural analyses of death domains and their interactions. Apoptosis 2011, 16, 209–220. [Google Scholar]
- Park, H.H.; Logette, E.; Rauser, S.; Cuenin, S.; Walz, T.; Tschopp, J.; Wu, H. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell 2007, 128, 533–546. [Google Scholar]
- Park, H.H.; Lo, Y.C.; Lin, S.C.; Wang, L.; Yang, J.K.; Wu, H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Ann. Rev. Immunol 2007, 25, 561–586. [Google Scholar]
- Lavrik, I.; Golks, A.; Krammer, P.H. Death receptor signaling. J. Cell Sci 2005, 118, 265–267. [Google Scholar]
- Peter, M.E.; Krammer, P.H. The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ 2003, 10, 26–35. [Google Scholar]
- Medema, J.P.; Scaffidi, C.; Kischkel, F.C.; Shevchenko, A.; Mann, M.; Krammer, P.H.; Peter, M.E. FLICE is activated by association with the CD95 death-inducing signaling complex (DISC). EMBO J 1997, 16, 2794–2804. [Google Scholar]
- Acehan, D.; Jiang, X.; Morgan, D.G.; Heuser, J.E.; Wang, X.; Akey, C.W. Three-dimensional structure of the apoptosome: Implications for assembly, procaspase-9 binding, and activation. Mol. Cell 2002, 9, 423–432. [Google Scholar]
- Shiozaki, E.N.; Chai, J.; Shi, Y. Oligomerization and activation of caspase-9, induced by Apaf-1 CARD. Proc. Natl. Acad. Sci. USA 2002, 99, 4197–4202. [Google Scholar]
- Jiang, X.; Wang, X. Cytochrome c promotes caspase-9 activation by inducing nucleotide binding to Apaf-1. J. Biol. Chem 2000, 275, 31199–31203. [Google Scholar]
- Lassus, P.; Opitz-Araya, X.; Lazebnik, Y. Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science 2002, 297, 1352–1354. [Google Scholar]
- Tinel, A.; Tschopp, J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 2004, 304, 843–846. [Google Scholar]
- Jang, T.H.; Zheng, C.; Wu, H.; Jeon, J.H.; Park, H.H. In vitro reconstitution of the interactions in the PIDDosome. Apoptosis 2010, 15, 1444–1452. [Google Scholar]
- Jang, T.H.; Bae, J.Y.; Park, O.K.; Kim, J.H.; Cho, K.H.; Jeon, J.H.; Park, H.H. Identification and analysis of dominant negative mutants of RAIDD and PIDD. Biochim. Biophys. Acta 2010, 1804, 1557–1563. [Google Scholar]
- Salvesen, G.S. Caspases and apoptosis. Essays Biochem 2002, 38, 9–19. [Google Scholar]
- Riedl, S.J.; Shi, Y. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol 2004, 5, 897–907. [Google Scholar]
- Chao, Y.; Shiozaki, E.N.; Srinivasula, S.M.; Rigotti, D.J.; Fairman, R.; Shi, Y. Engineering a dimeric caspase-9: A re-evaluation of the induced proximity model for caspase activation. PLoS Biol 2005, 3. [Google Scholar] [CrossRef] [Green Version]
- Yuan, S.; Yu, X.; Topf, M.; Ludtke, S.J.; Wang, X.; Akey, C.W. Structure of an apoptosome-procaspase-9 CARD complex. Structure 2010, 18, 571–583. [Google Scholar]
- Denault, J.B.; Salvesen, G.S. Caspases: Keys in the ignition of cell death. Chem. Rev 2002, 102, 4489–4500. [Google Scholar]
- Salvesen, G.S.; Dixit, V.M. Caspase activation: the induced-proximity model. Proc. Natl. Acad. Sci. USA 1999, 96, 10964–10967. [Google Scholar]
- Reed, J.C.; Doctor, K.S.; Godzik, A. The domains of apoptosis: A genomics perspective. Sci. STKE 2004. [Google Scholar] [CrossRef]
- Huang, B.; Eberstadt, M.; Olejniczak, E.T.; Meadows, R.P.; Fesik, S.W. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature 1996, 384, 638–641. [Google Scholar]
- Eberstadt, M.; Huang, B.; Chen, Z.; Meadows, R.P.; Ng, S.-C.; Zheng, L.; Lenardo, M.J.; Fesik, S.W. NMR structure and mutagenesis of the FADD (Mort1) death-effector domain. Nature 1998, 392, 941–945. [Google Scholar]
- Chou, J.J.; Matsuo, H.; Duan, H.; Wagner, G. Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment. Cell 1998, 94, 171–180. [Google Scholar]
- Hiller, S.; Kohl, A.; Fiorito, F.; Herrmann, T.; Wider, G.; Tschopp, J.; Grutter, M.G.; Wuthrich, K. NMR structure of the apoptosis- and inflammation-related NALP1 pyrin domain. Structure 2003, 11, 1199–1205. [Google Scholar]
- Wajant, H. The Fas signaling pathway: More than a paradigm. Science 2002, 296, 1635–1636. [Google Scholar]
- Li, P.; Nijhawan, D.; Budihardjo, I.; Srinivasula, S.M.; Ahmad, M.; Alnemri, E.S.; Wang, X. Cytochrome c and dATP-dependent formation of Apaf-1/Caspase-9 complex initiates an apoptotic protease cascade. Cell 1997, 91, 479–489. [Google Scholar]
- Carrington, P.E.; Sandu, C.; Wei, Y.; Hill, J.M.; Morisawa, G.; Huang, T.; Gavathiotis, E.; Werner, M.H. The structure of FADD and its mode of interaction with procaspase-8. Mol. Cell 2006, 22, 599–610. [Google Scholar]
- Scott, F.L.; Stec, B.; Pop, C.; Dobaczewska, M.K.; Lee, J.J.; Monosov, E.; Robinson, H.; Salvesen, G.S.; Schwarzenbacher, R.; Riedl, S.J. The Fas-FADD death domain complex structure unravels signalling by receptor clustering. Nature 2009, 457, 1019–1022. [Google Scholar]
- Wang, L.; Yang, J.K.; Kabaleeswaran, V.; Rice, A.J.; Cruz, A.C.; Park, A.Y.; Yin, Q.; Damko, E.; Jang, S.B.; Raunser, S.; et al. The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations. Nat. Struct. Mol. Biol 2010, 17, 1324–1329. [Google Scholar]
- Adams, J.M.; Cory, S. Apoptosomes: engines for caspase activation. Curr. Opin. Cell Biol 2002, 14, 715–720. [Google Scholar]
- Qin, H.; Srinivasula, S.M.; Wu, G.; Fernandes-Alnemri, T.; Alnemri, E.S.; Shi, Y. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature 1999, 399, 549–557. [Google Scholar]
- Rodriguez, J.; Lazebnik, Y. Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev 1999, 13, 3179–3184. [Google Scholar]
- Yuan, S.; Yu, X.; Topf, M.; Dorstyn, L.; Kumar, S.; Ludtke, S.J.; Akey, C.W. Structure of the Drosophila apoptosome at 6.9 Å structure. Structure 2011, 19, 128–140. [Google Scholar]
- Qi, S.; Pang, Y.; Hu, Q.; Liu, Q.; Li, H.; Zhou, Y.; He, T.; Liang, Q.; Liu, Y.; Yuan, X.; et al. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell 2010, 141, 446–457. [Google Scholar]
- Guo, Y.; Srinivasula, S.M.; Druilhe, A.; Fernandes-Alnemri, T.; Alnemri, E.S. Caspase-2 induces apoptosis by releasing proapoptotic proteins from mitochondria. J. Biol. Chem 2002, 277, 13430–13437. [Google Scholar]
- Janssens, S.; Tinel, A.; Lippens, S.; Tschopp, J. PIDD mediates NF-kappaB activation in response to DNA damage. Cell 2005, 123, 1079–1092. [Google Scholar]
- Pick, R.; Badura, S.; Bosser, S.; Zornig, M. Upon intracellular processing, the C-terminal death domain-containing fragment of the p53-inducible PIDD/LRDD protein translocates to the nucleoli and interacts with nucleolin. Biochem. Biophys. Res. Commun 2006, 349, 1329–1338. [Google Scholar]
- Tinel, A.; Janssens, S.; Lippens, S.; Cuenin, S.; Logette, E.; Jaccard, B.; Quadroni, M.; Tschopp, J. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-kappaB pathway. EMBO J 2007, 26, 197–208. [Google Scholar]
- Bae, J.Y.; Park, H.H. Crystal structure of NALP3 PYD and its implication in inflammasome assembly. J. Biol. Chem 2011, 286, 39528–39536. [Google Scholar]
- Kwon, D.; Yoon, J.W.; Shin, S.; Jang, T.; Kim, H.; So, I.; Jeon, J.; Park, H.H. A comprehensive manually curated protein-protein interaction database for the death domain superfamily. Nucleic Acids Res 2012, 40, 331–336. [Google Scholar]


© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Park, H.H. Structural Features of Caspase-Activating Complexes. Int. J. Mol. Sci. 2012, 13, 4807-4818. https://doi.org/10.3390/ijms13044807
Park HH. Structural Features of Caspase-Activating Complexes. International Journal of Molecular Sciences. 2012; 13(4):4807-4818. https://doi.org/10.3390/ijms13044807
Chicago/Turabian StylePark, Hyun Ho. 2012. "Structural Features of Caspase-Activating Complexes" International Journal of Molecular Sciences 13, no. 4: 4807-4818. https://doi.org/10.3390/ijms13044807



