Extreme Multiple Reticulate Origins of the Pteris cadieri Complex (Pteridaceae)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Results
2.1.1. Morphological Discrimination
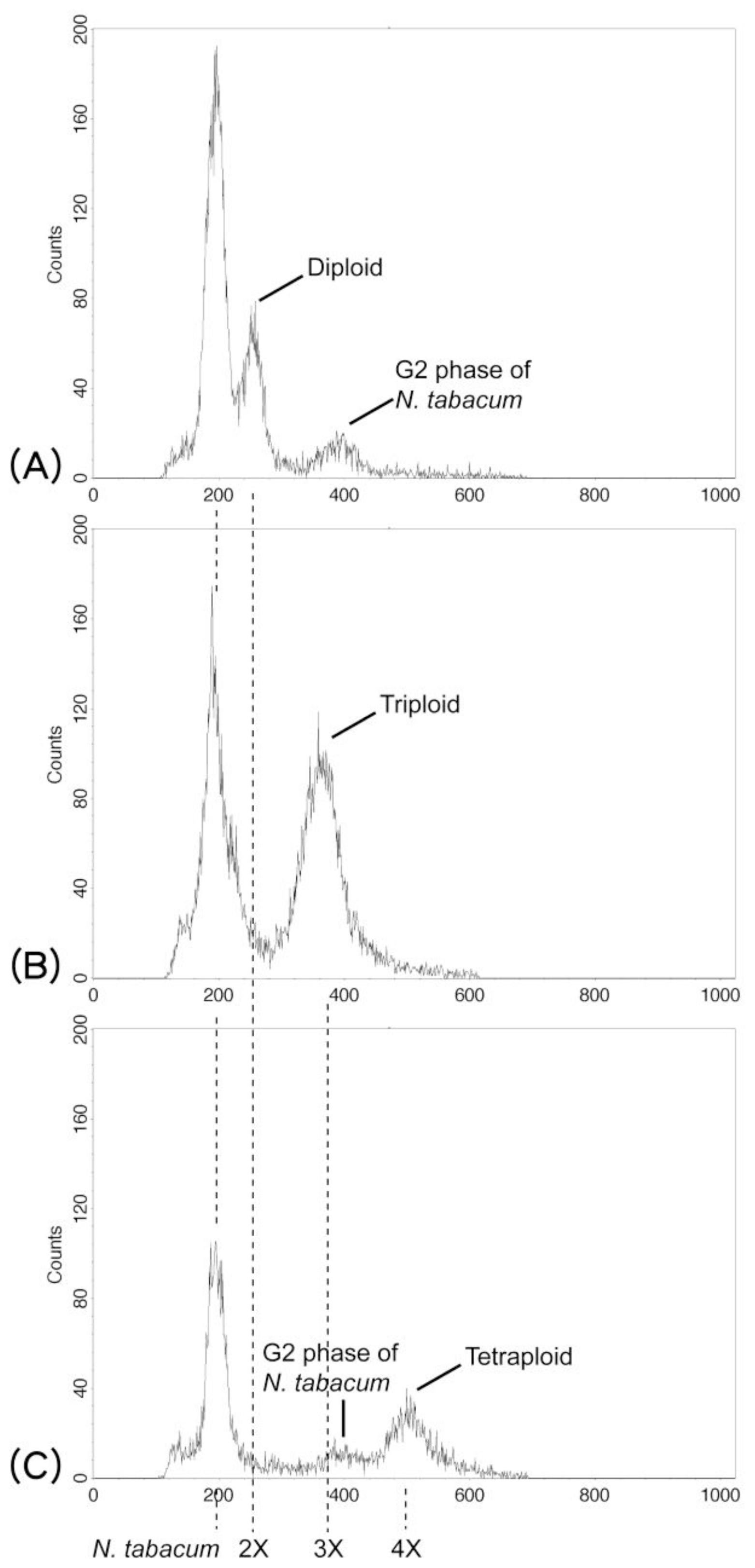
2.1.2. Cytotypes, Phenology, and Reproductive Systems
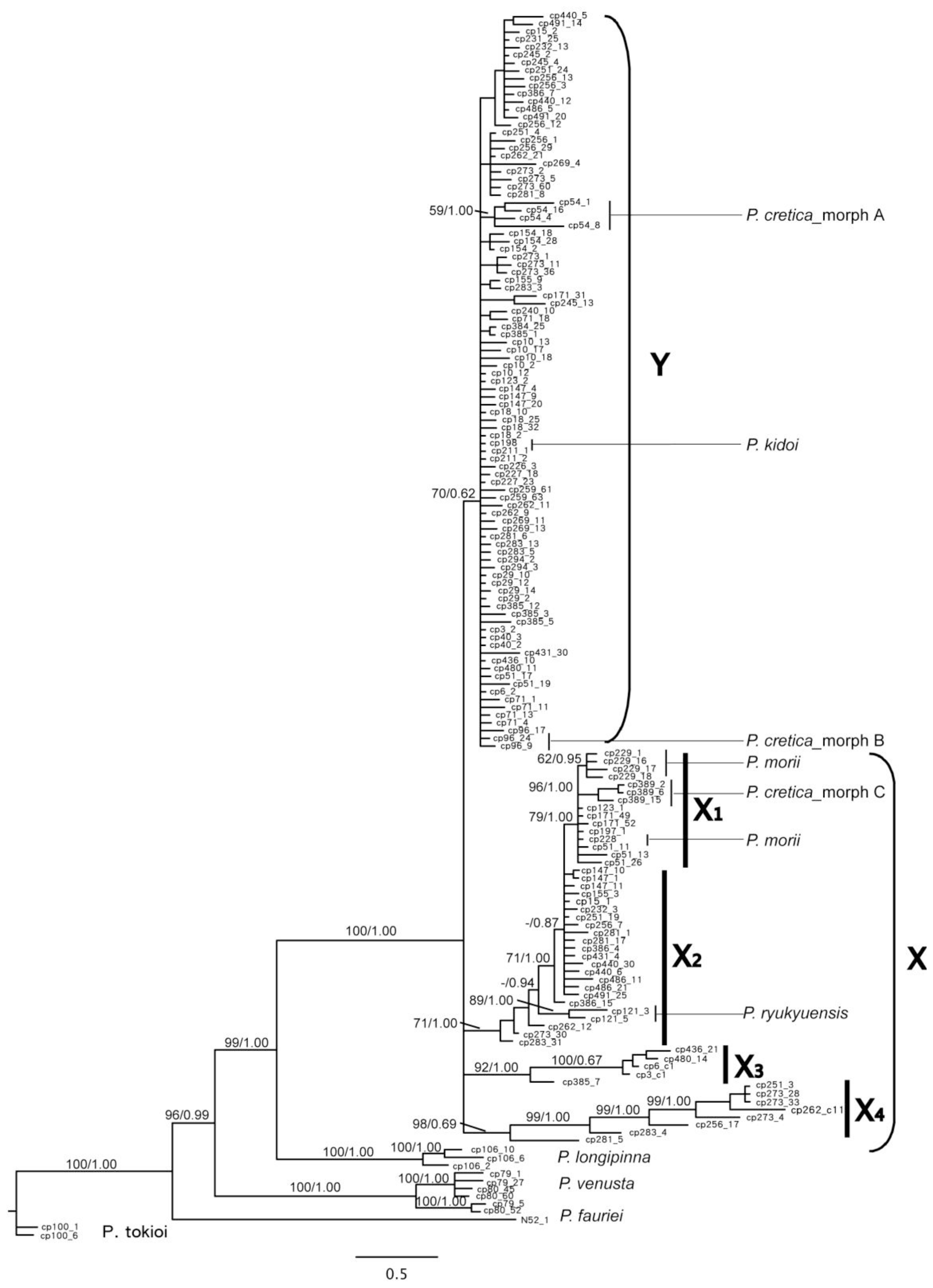
2.1.3. Phylogenetic Resolution of Chloroplast DNA Data
2.1.4. Phylogenetic Resolution of PgiC Gene Data
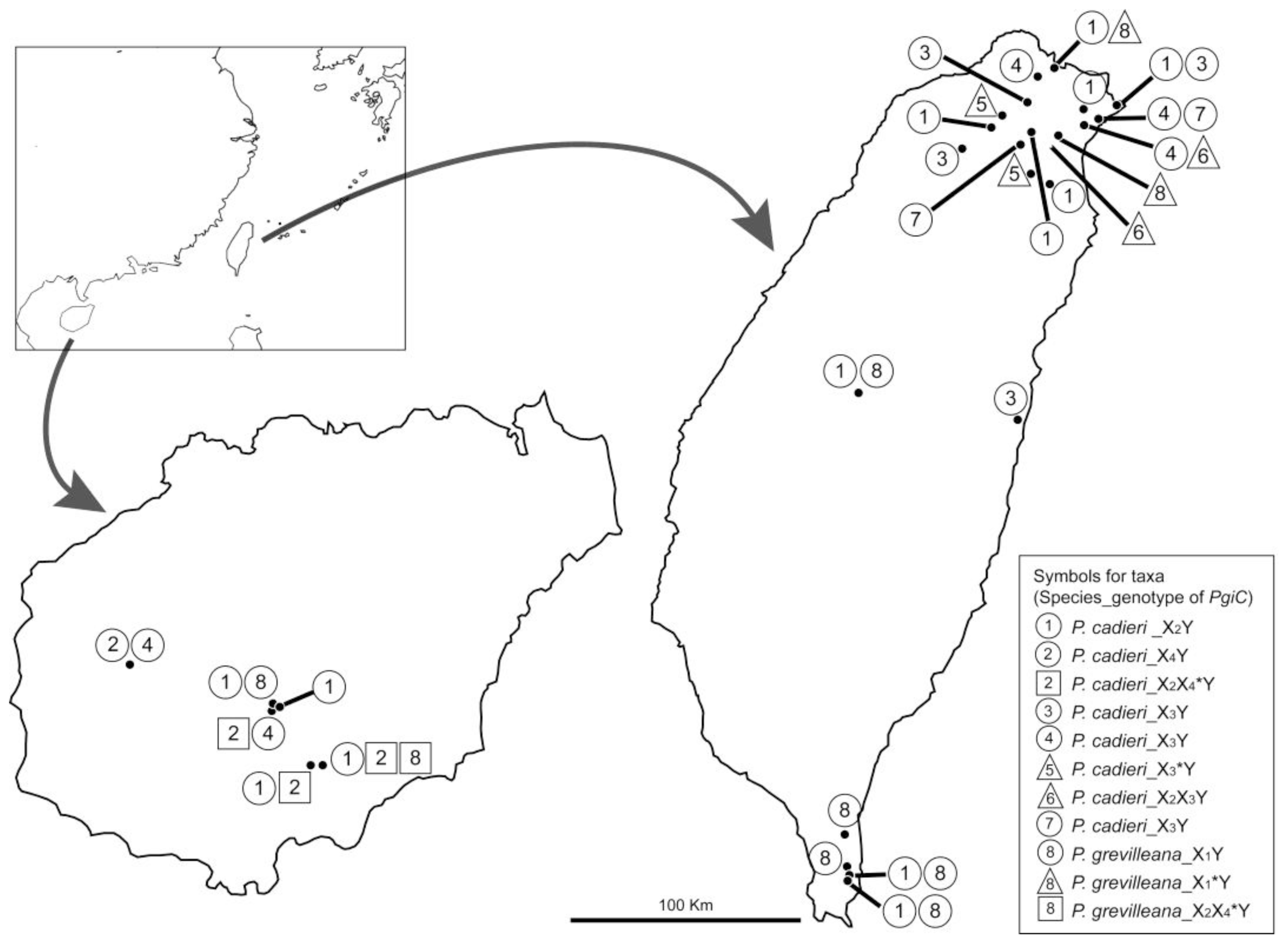
2.1.5. Divergence in Different Geographical Areas and Phenology
2.2. Discussion
2.2.1. Hybrid Origin
2.2.2. Parental Species?
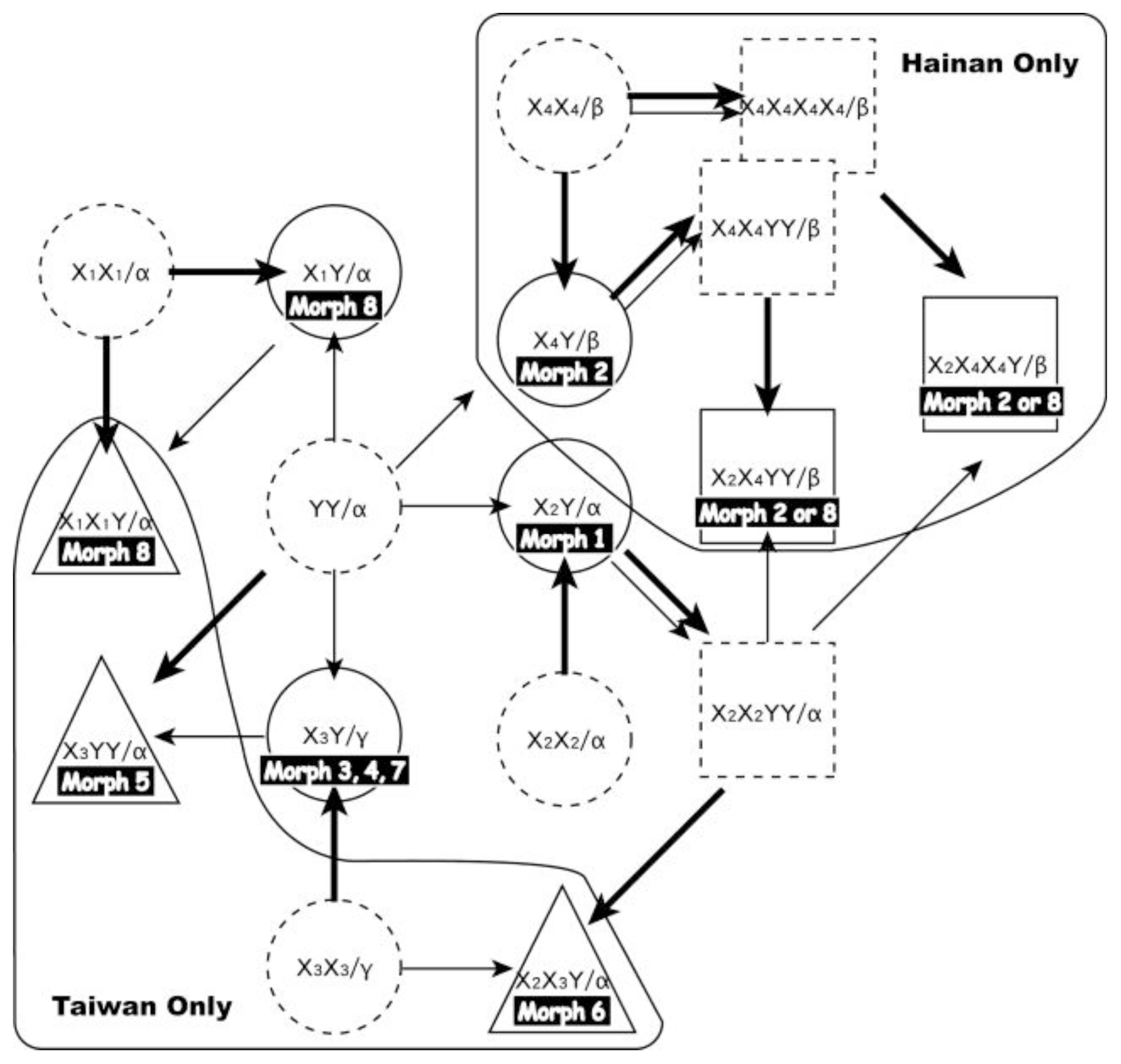
2.2.3. Reticulate Evolution of the Pteris cadieri Complex
2.2.4. Multiple Origins Supported by Genotypic, Geographic, and Phenological Divergence
3. Experimental Section
3.1. Sampling and Phenological Studies
3.2. Ploidy Analysis and Reproductive Systems
3.3. Molecular Methods
3.4. Phylogenetic Analysis
4. Conclusions
Supplementary Information
ijms-13-04523-s001.pdfAcknowledgments
References
- Hewitt, G.M. Hybrid zones—Natural laboratories for evolutionary studies. Trends Ecol. Evol 1988, 3, 158–167. [Google Scholar]
- Harrison, R.G. Hybrid Zones and the Evolutionary Process; Oxford University Press: New York NY, USA, 1993; p. 364. [Google Scholar]
- Van Droogenbroeck, B.; Kyndt, T.; Romeijn-Peeters, E.; van Thuyne, W.; Goetghebeur, P.; Romero-Motochi, J.P.; Gheysen, G. Evidence of natural hybridization and introgression between Vasconcellea species (Caricaceae) from southern Ecuador revealed by chloroplast, mitochondrial and nuclear DNA markers. Ann. Bot 2006, 97, 793–805. [Google Scholar]
- Kelly, L.J.; Leitch, A.R.; Clarkson, J.J.; Hunter, R.B.; Knapp, S.; Chase, M.W. Intragenic recombination events and evidence for hybrid speciation in Nicotiana (Solanaceae). Mol. Biol. Evol 2010, 27, 781–799. [Google Scholar]
- Lihová, J.; Kučera, J.; Perný, M.; Marhold, K. Hybridization between two polyploid Cardamine (Brassicaceae) species in north-western Spain: Discordance between morphological and genetic variation patterns. Ann. Bot 2007, 99, 1083–1096. [Google Scholar]
- Shepherd, L.D.; Perrie, L.R.; Brownsey, P.J. Low-copy nuclear DNA sequences reveal a predominance of allopolyploids in a New Zealand Asplenium fern complex. Mol. Biol. Evol 2008, 49, 240–248. [Google Scholar]
- Sciarretta, K.L.; Arbuckle, E.P.; Haufler, C.H.; Werth, C.R. Patterns of genetic variation in southern Appalachian populations of Athyrium filix-femina var. asplenioides (Dryopteridaceae). Int. J. Plant Sci 2005, 166, 761–780. [Google Scholar]
- Kentner, E.K.; Mesler, M.R. Evidence for natural selection in a fern hybrid zone. Am. J. Bot 2000, 87, 1168–1174. [Google Scholar]
- Grusz, A.L.; Windham, M.D.; Pryer, K.M. Deciphering the origins of apomictic polyploids in the Cheilanthes yavapensis complex (Pteridaceae). Am. J. Bot 2009, 96, 1636–1645. [Google Scholar]
- Schuettpelz, E.; Grusz, A.L.; Windham, M.D.; Pryer, K.M. The utility of nuclear gapCp in resolving polyploid fern origins. Syst. Bot 2008, 33, 621–629. [Google Scholar]
- Ebihara, A.; Ishikawa, H.; Matsumoto, S.; Lin, S.J.; Iwatsuki, K.; Takamiya, M.; Watano, Y.; Ito, M. Nuclear DNA, chloroplast DNA, and ploidy analysis clarified biological complexity of the Vandenboschia radicans complex (Hymenophyllaceae) in Japan and adjacent areas. Am. J. Bot 2005, 92, 1535–1547. [Google Scholar]
- Ootsuki, R.; Sato, H.; Nakato, N.; Murakami, N. Evidence of genetic segregation in the apogamous fern species Cyrtomium fortunei (Dryopteridaceae). J. Plant Res 2012. [Google Scholar] [CrossRef]
- Lovis, J.D. Evolutionary patterns and processes in ferns. Adv. Bot. Res 1977, 4, 229–414. [Google Scholar]
- Walker, T.G. The Cytogenetics of Ferns. In The Experimental Biology of Ferns; Dyer, A.F., Ed.; Academic Press: London, UK, 1979; pp. 87–132. [Google Scholar]
- Kato, M.; Nakato, N.; Cheng, X.; Iwatsuki, K. Cytotaxonomic study of ferns of Yunnan, southwestern China. Bot. Mag. Tokyo 1992, 105, 105–124. [Google Scholar]
- Simizu, H. Notes on the noteworthy ferns from Taiwan. Trans. Nat. Hist. Soc. Taiwan 1938, 28, 238–246. [Google Scholar]
- Shieh, W.C. Pteridaceae. In Flora of Taiwan, 1st ed; Li, H.-L., Liu, T.-C., Koyama, T., DeVol, C.E., Eds.; Epoch Publishing: Taipei, Taiwan, 1975; Volume 1, pp. 281–301. [Google Scholar]
- Ching, R.-C.; Wang, C.H. Materials for the pteridophytic flora of Hainan. Acta Phytotax. Sin 1959, 8, 125–171. [Google Scholar]
- Wu, S.-W. Pteridaceae. In Flor. Reipubl. Pop. Sinicae; Ching, R.-C., Shing, K.-H., Eds.; Science Press: Beijing, China, 1990; Volume 3, pp. 15–91. [Google Scholar]
- Walker, E.H. Pteridaceae. In Flora of Okinawa and the southern Ryukyu Islands; Smithsonian Institution Press: Washington, DC, USA, 1976. [Google Scholar]
- Christ, M.H. Flilices cadierianae. J. Bot. (Morot) 1905, 19, 58–79. [Google Scholar]
- Kuo, C.-M.; Yu, H.-T. Investigation on the Pteridophyta of Kenting National Park; Kenting National Park Administration Office: Hengchun Township, Taiwan, 1986; p. 114. [Google Scholar]
- Wagner, W.H.J. Venuloid idioblasts in Pteris and their systematic implication. Acta Phytotax. Geobot 1978, 29, 33–40. [Google Scholar]
- Nakaike, T. New Flora of Japan: Pteridophyta. Revised & Enlarged; Shibundo Company: Tokyo, Japan, 1992. [Google Scholar]
- Chao, Y.S.; Liu, H.Y.; Huang, Y.M.; Chiou, W.L. Reproductive traits of Pteris cadieri and P. grevilleana in Taiwan: Implications for their hybrid origins. Bot. Stud 2010, 51, 209–216. [Google Scholar]
- Levin, D.A. The Origin, Expansion, and Demise of Plant Species; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Soltis, D.E.; Soltis, P.S. Molecular data and the dynamic nature of polyploidy. Crit. Rev. Plant Sci 1993, 12, 243–273. [Google Scholar]
- Schwarzbach, A.E.; Rieseberg, L.H. Likely multiple origins of a diploid hybrid sunflower species. Mol. Ecol 2002, 11, 1703–1715. [Google Scholar]
- Soltis, D.E.; Soltis, P.S.; Bennett, M.D.; Leitch, I.J. Evolution of genome size in the angiosperms. Am. J. Bot 2003, 90, 1596–1603. [Google Scholar]
- Chat, J.; Jauregui, B.; Petit, R.J.; Nadot, S. Reticulate evolution in kiwifruit (Actinidia, actinidiaceae) identified by comparing their maternal and paternal phylogenies. Am. J. Bot 2004, 91, 736–747. [Google Scholar]
- Yang, W.; Glover, B.J.; Rao, G.-Y.; Yang, J. Molecular evidence for multiple polyploidization and lineage recombination in the Chrysanthemum indicum polyploid complex (Asteraceae). New Phytol 2006, 171, 875–886. [Google Scholar]
- Guggisberg, A.; Bretagnolle, F.; Mansion, G. Allopolyploid origin of the mediterranean endemic, Centaurium bianoris (Gentianaceae), inferred by molecular markers. Syst. Bot 2009, 31, 368–379. [Google Scholar]
- Nelson, A.D.; Elisens, W.J. Polyploid evolution and biogeography in Chelone (Scrophulariaceae): Morphological and isozyme evidence. Am. J. Bot 1999, 86, 1487–1501. [Google Scholar]
- Noyes, R.D. Intraspecific nuclear ribosomal DNA divergence and reticulation in sexual diploid Erigeron strigosus (Asteraceae). Am. J. Bot 2006, 93, 470–479. [Google Scholar]
- Watanabe, K.; Ohi-Toma, T.; Murata, J. Multiple hybridization in the Aristolochia kaempferi group (Aristolochiaceae): Evidence from reproductive isolation and molecular phylogeny. Am. J. Bot 2008, 95, 885–896. [Google Scholar]
- Baldwin, B.G.; Sanderson, M.J.; Porter, J.M.; Wojciechowski, M.F.; Campbell, C.S.; Donoghue, M.J. The ITS region of nuclear ribosomal DNA: A valuable source of evidence on angiosperm phylogeny. Ann. Mo. Bot. Gard 1995, 82, 247–277. [Google Scholar]
- Álvarez, I.; Wendel, J.F. Ribosomal its sequences and plant phylogenetic inference. Mol. Phylogenet. Evol 2003, 29, 417–434. [Google Scholar]
- Friar, E.A.; Prince, L.M.; Cruse-Sanders, J.M.; McGlaughlin, M.E.; Butterworth, C.A.; Baldwin, B.G. Hybrid origin and genomic mosaicism of Dubautia scabra (Hawaiian silversword alliance; Asteraceae, Madiinae). Syst. Bot 2008, 33, 589–597. [Google Scholar]
- Hoot, S.B.; Taylor, W.C. The utility of nuclear ITS, a leafy homolog intron, and chloroplast atpB-rbcL spacer region data in phylogenetic analyses and species delimitation in Isoëtes. Am. Fern J 2001, 91, 166–177. [Google Scholar]
- Adjie, B.; Masuyama, S.; Ishikawa, H.; Watano, Y. Independent origins of tetraploid cryptic species in the fern Ceratopteris thalictroides. J. Plant Res 2007, 120, 129–138. [Google Scholar]
- Ishikawa, H.; Watano, Y.; Kano, K.; Ito, M.; Kurit, S. Development of primer sets for PCR amplification of the PgiC gene in ferns. J. Plant Res 2002, 115, 65–70. [Google Scholar]
- Chang, H.-M.; Chiou, W.-L.; Wang, J.-C. Molecular evidence for genetic heterogeneity and the hybrid origin of Acrorumohra subreflexipinna from Taiwan. Am. Fern J 2009, 99, 61–77. [Google Scholar]
- Dong, S.Y.; Chen, Z.C.; Zhang, X.C. A survey of the pteridophytes of Hainan Island, China. Living For 2003, 6, 36–37. [Google Scholar]
- Dong, S.Y.; Chen, Z.C.; Zhang, X.C. Additions to the pteridophyte flora of Hainan Island. Bull. Bot. Res 2004, 24, 45–48. [Google Scholar]
- Johnston, J.S.; Bennett, M.D.; Rayburn, A.L.; Galbraith, D.W.; Price, H.J. Reference standards for determination of DNA content of plant nuclei. Am. J. Bot 1999, 86, 609–613. [Google Scholar]
- Wolf, P.G. Evaluation of atpB nucleotide sequences for phylogenetic studies of ferns and other pteridophytes. Am. J. Bot 1997, 84, 1429–1440. [Google Scholar]
- Pryer, K.M.; Schuettpelz, E.; Wolf, P.G.; Schneider, H.; Smith, A.R.; Cranfill, R. Phylogeny and evolution of ferns (monilophytes) with a focus on the early leptosporangiate divergences. Am. J. Bot 2004, 91, 1582–1598. [Google Scholar]
- Wolf, P.G.; Soltis, P.S.; Soltis, D.E. Phylogenetic relationships of dennstaedtioid ferns: Evidence from rbcL sequences. Mol. Phylogenet. Evol 1994, 3, 383–392. [Google Scholar]
- Pryer, K.M.; Smith, A.R.; Hunt, J.S.; Dubuisson, J.-Y. RbcL data reveal two monophyletic groups of filmy ferns (Filicopsida: Hymenophyllaceae). Am. J. Bot 2001, 88, 1118–1130. [Google Scholar]
- Wagner, A.; Blackstone, N.; Cartwright, P.; Dick, M.; Misof, B.; Snow, P.; Wagner, G.P.; Bartels, J.; Murtha, M.; Pendleton, J. Surveys of gene families using polymerase chain reaction: PCR selection and PCR drift. Syst. Biol 1994, 43, 250–261. [Google Scholar]
- Harris, J.L.; Ingram, R. Chloroplast DNA and biosystematics: The effects of intraspecific diversity and plasmid transmission. Taxon 1991, 40, 393–421. [Google Scholar]
- Walker, T.G. Cytology and evolution in the fern genus Pteris l. Evolution 1962, 16, 17–43. [Google Scholar]
- Manton, I. Problems of Cytology and Evolution in the Pteridophyta; Cambridge University Press: New York, NY, USA, 1950; p. 316. [Google Scholar]
- Suzuki, T.; Iwatsuki, K. Genetic variation in apogamous fern Pteris cretica l. In Japan. Heredity 1990, 65, 221–227. [Google Scholar]
- Nakaike, T. Index of Hybrid Ferns in Japan; University of Tokyo: Tokyo, Japan, 1970. [Google Scholar]
- Beck, J.B.; Windham, M.D.; Yatskievych, G.; Pryer, K.M. A diploids-first approach to species delimitation and interpreting polyploid evolution in the fern genus Astrolepis (Pteridaceae). Syst. Bot 2010, 35, 223–234. [Google Scholar]
- Haufler, C.H.; Windham, M.D.; Britton, D.M.; Robinson, S.J. Triploidy and its evolutionary significance in Cystopteris protrusa. Can. J. Bot 1985, 63, 1855–1863. [Google Scholar]
- Lin, S.-J.; Kato, M.; Iwatsuki, K. Electrophoretic variation of apogamous Dryopteris varia group (Drypteridaceae). J. Plant Res 1995, 108, 451–456. [Google Scholar]
- Lin, S.-J.; Kato, M.; Iwatsuki, K. Diploid and triploid offspring of triploid agamosporous fern Dryopteris pacifica. Bot. Mag. Tokyo 1992, 105, 443–452. [Google Scholar]
- Huang, Y.-M.; Chou, H.-M.; Wang, J.-C.; Chiou, W.-L. The distribution and habitats of the Pteris fauriei complex. Taiwania 2007, 52, 49–58. [Google Scholar]
- Hickok, L.G.; Klekowski, E.J., Jr. Inchoate speciation in Ceratopteris: An analysis of the synthesized hybrid C. richardii × C. pteridoides. Evolution 1974, 28, 439–446. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull 1987, 19, 11–15. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997, 24, 4876–4882. [Google Scholar]
- Swofford, D.L. Paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods), version 4; Sinauer: Sunderland, MA, USA, 2002. [Google Scholar]
- Farris, J.S.; Kallersjo, M.; Kluge, A.G.; Bult, C. Testing significance of incongruence. Cladistics 1995, 10, 315–320. [Google Scholar]
- Simmons, M.P.; Ochoterena, H. Gaps as characters in sequence-based phylogenetic analyses. Syst. Biol 2000, 49, 369–381. [Google Scholar]
- Young, N.D.; Healy, J. Gapcoder automates the use of indel characters in phylogenetic analysis. BMC Bioinforma 2003, 4, 1–6. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar]
- Zwickl, D.J. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets under the Maximum Likelihood Criterion. Ph.D. Dissertation, University of Texas, Austin, TX, USA, 2006. [Google Scholar]
- Huelsenbeck, J.P.; Ronquist, F. Mrbayes: Bayesian inference of phylogeny. Bioinformatics 2001, 17, 754–755. [Google Scholar]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol 1993, 10, 512–526. [Google Scholar]
- Posada, D.; Crandall, K.A. Modeltest: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar]
- Rambaut, A.; Drummond, A.J. Tracer v1.5. 2009. Available online: http://tree.Bio.Ed.Ac.Uk/software/tracer/ accessed on 2 December 2009.
- Hörandl, E.; Stuessy, T.F. Paraphyletic groups as natural units of biological classification. Taxon 2010, 59, 1641–1653. [Google Scholar]
- Zhuang, Y.; Adams, K.L. Extensive allelic variation in gene expression in Populus F1 hybrids. Genetics 2007, 177, 1987–1996. [Google Scholar]
- Guo, M.; Rupe, M.A.; Zinselmeier, C.; Habben, J.; Bowen, B.A.; Smith, O.S. Allelic variation of gene expression in maize hybrids. Plant Cell 2004, 16, 1707–1716. [Google Scholar]
- Springer, N.M.; Stupar, R.M. Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell 2007, 19, 2391–2402. [Google Scholar]
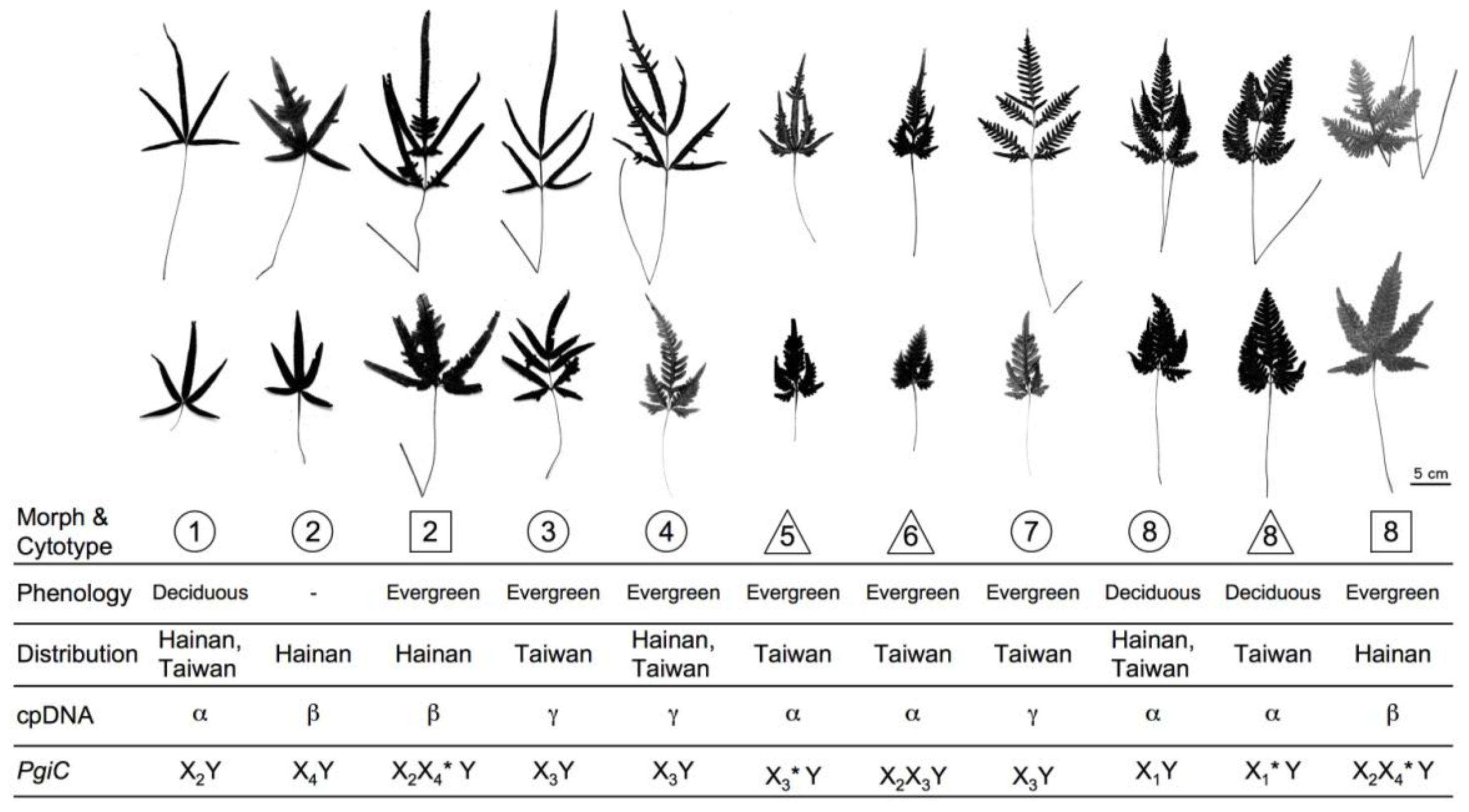

| Morph type | Ploidy level | Fertile blades | Sterile blades | Scales |
|---|---|---|---|---|
| Morph 1 | Diploid | Pedate or pinnate | Pedate, basal pinna straight or incurved | Wide and dark brown center |
| Morph 2 | Diploid, tetraploid | Pedate or pinnate, sometimes with digital projects | Pedate, basal pinna straight or incurved | Narrow and dark brown center |
| Morph 3 | Diploid | Pinnate, sometimes with digital projects | Pinnate, sometimes with digital projects, basal pinna straight | Wide and dark brown center |
| Morph 4 | Diploid | Pinnate, sometimes with digital projects | Bipinnatifid, basal pinna straight | Wide and dark brown center |
| Morph 5 | Triploid | Pedate, sometimes with digital projects | Bipinnatifid, basal pinna incurved | Wide and dark brown center |
| Morph 6 | Triploid | Irregularly bipinnatifid | Bipinnatifid, basal pinna incurved | Wide and dark brown center |
| Morph 7 | Diploid | Almost regularly bipinnatifid | Bipinnatifd, basal pinna straight | Wide and dark brown center |
| Morph 8 | Diploid, triploid, tetraploid | Bipinnatifid | Bipinnatifid, basal pinna straight or incurved | Wide and dark brown center |
| Primer for Chloroplast DNA | ||
|---|---|---|
| Primer | Primer sequence (5′→3′) | Origin |
| atpB_672 | CAC TSA GAG GRG CTC CCG TAT CAA | [46] |
| rbcL_r49R | CAC CAG CTT TGA ATC CAA CAC TTG C | [46] |
| atpB 493F | CGA CGA TAC GGR GCC AAA AGA TCC | [47] |
| rbcL r158R | AAG ATT CCG CAG CTA CTG CAG CTC C | [47] |
| rbcL F1F | ATG TCA CCA CAA ACA GAA ACT AAA GCA AGT | [48] |
| rbcL F1379R | TCA CAA GCA GCA GCT AGT TCA GGA CTC | [49] |
| rbcL_PF | TAA GTA TCG TGY GGA GGT TRA ATC A | This study |
| Primer for PgiC | ||
| Primer | Primer sequence (5′→3′) | Origin |
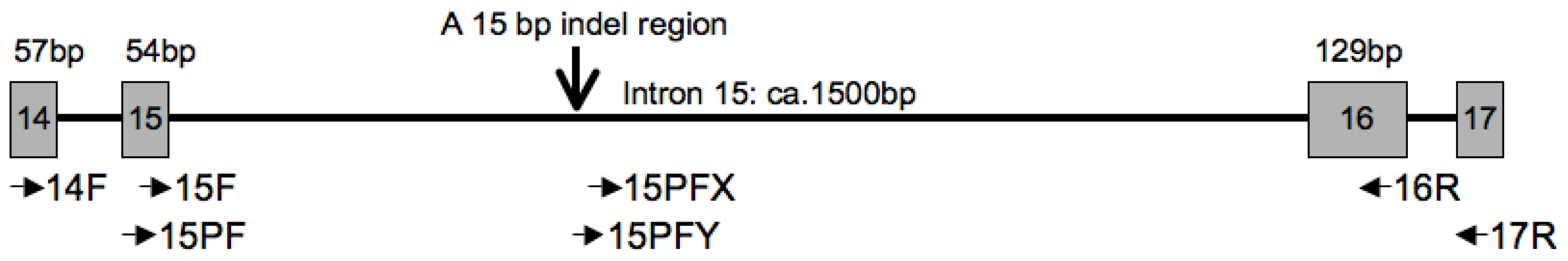
| 14F | GTG CTT CTG GGT CTT TTG AGT G | [41] |
| 16R | GTT GTC CAT TAG TTC CAG GTT CCC C | [41] |
| 15PF | CAAATCCTTTCTTGCAATAGGC | This study |
| 17R | GAA ATCAC ATGGA ATAAC ACGTCC | This study |
| 15PFX | CAAGT ATACC TCTTC TTGAC AG | This study |
| 15PFY | CAG CAA GTA TAA CAA AAA CTC GC | This study |
| atpB-rbcL spacer & rbcL gene | PgiC gene (primers 15PF and 17R) | PgiC gene (primers 15PFX/15PFY and 17R) | |
|---|---|---|---|
| No. sequence | 57 | 151 | 191 |
| Aligned length (bp) | 2126 | 1794 | 1090 |
| Characters included (bp) | 2091–2119 | 1431–1705 | 786–1050 |
| Variable characters (bp) | 227 | 559 | 399 |
| Parsimony informative characters (bp) | 98 | 306 (50 binary characters produced by coding indels) | 200 (41 binary characters produced by coding indels) |
| Obtained trees | 322 | 4128 | 1145 |
| Tree length | 300 | 774 | 578 |
| CI | 0.777 | 0.764 | 0.739 |
| RI | 0.930 | 0.929 | 0.937 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chao, Y.-S.; Dong, S.-Y.; Chiang, Y.-C.; Liu, H.-Y.; Chiou, W.-L. Extreme Multiple Reticulate Origins of the Pteris cadieri Complex (Pteridaceae). Int. J. Mol. Sci. 2012, 13, 4523-4544. https://doi.org/10.3390/ijms13044523
Chao Y-S, Dong S-Y, Chiang Y-C, Liu H-Y, Chiou W-L. Extreme Multiple Reticulate Origins of the Pteris cadieri Complex (Pteridaceae). International Journal of Molecular Sciences. 2012; 13(4):4523-4544. https://doi.org/10.3390/ijms13044523
Chicago/Turabian StyleChao, Yi-Shan, Shi-Yong Dong, Yu-Chung Chiang, Ho-Yih Liu, and Wen-Liang Chiou. 2012. "Extreme Multiple Reticulate Origins of the Pteris cadieri Complex (Pteridaceae)" International Journal of Molecular Sciences 13, no. 4: 4523-4544. https://doi.org/10.3390/ijms13044523




