Characterization of Novel Di-, Tri-, and Tetranucleotide Microsatellite Primers Suitable for Genotyping Various Plant Pathogenic Fungi with Special Emphasis on Fusaria and Mycospherella graminicola
Abstract
:1. Introduction
2. Experimental Section
2.1. Culture of Fungal Species and DNA Extraction
2.2. Primer Design
2.3. Microsatellite Primed PCR
2.4. Visual Analysis of Banding Patterns and Gel Documentation
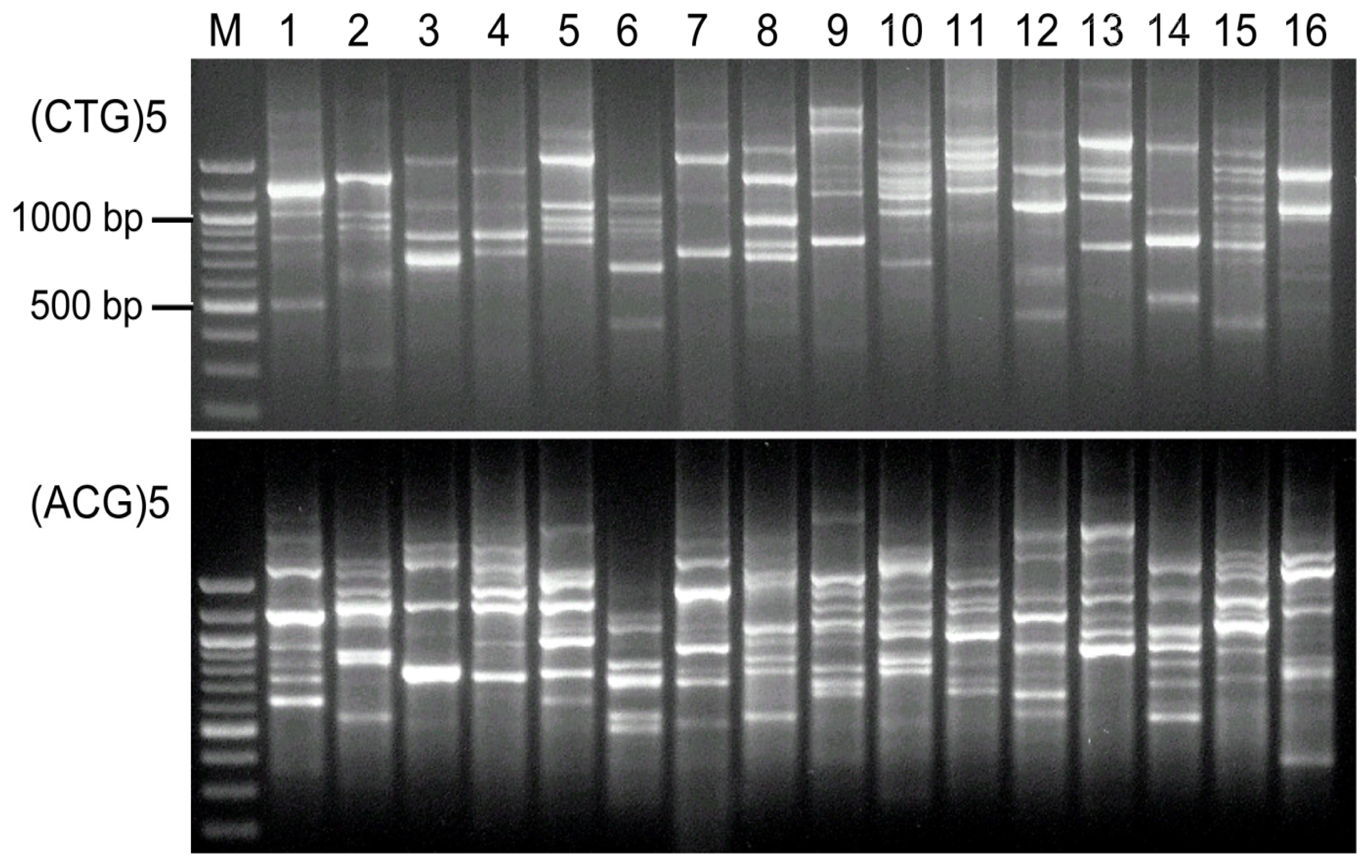
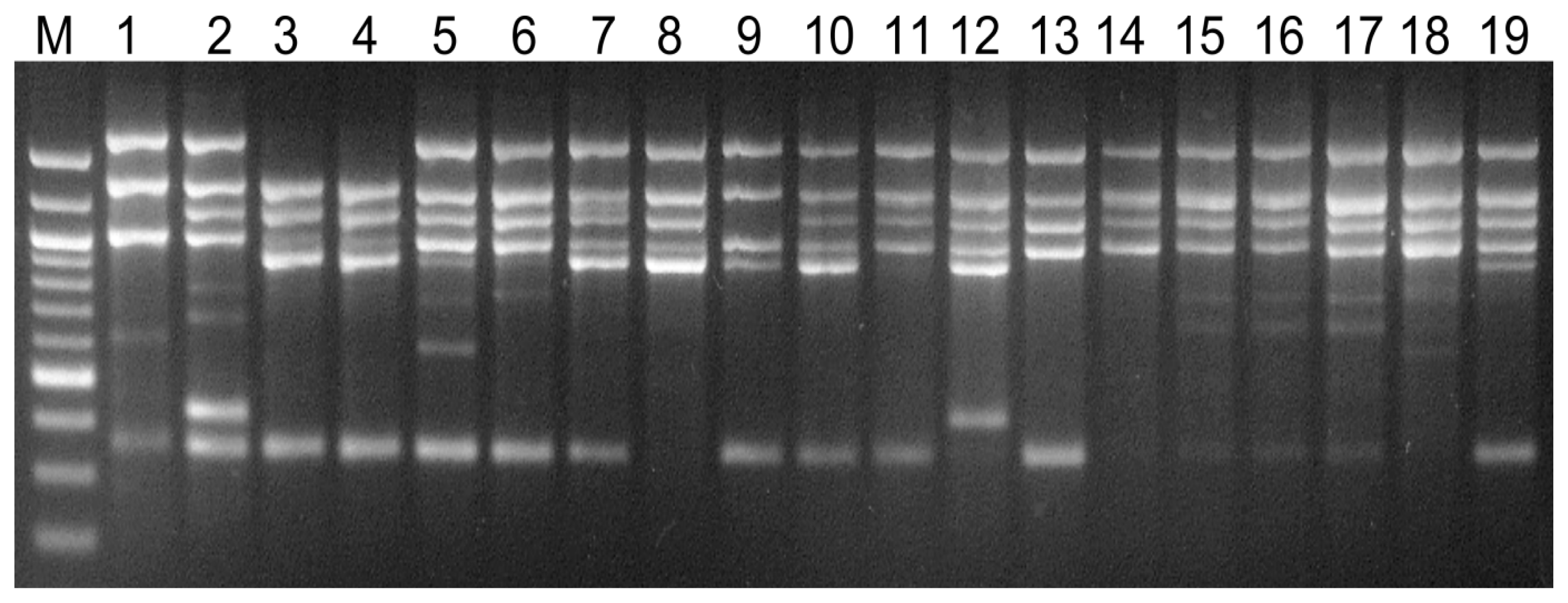
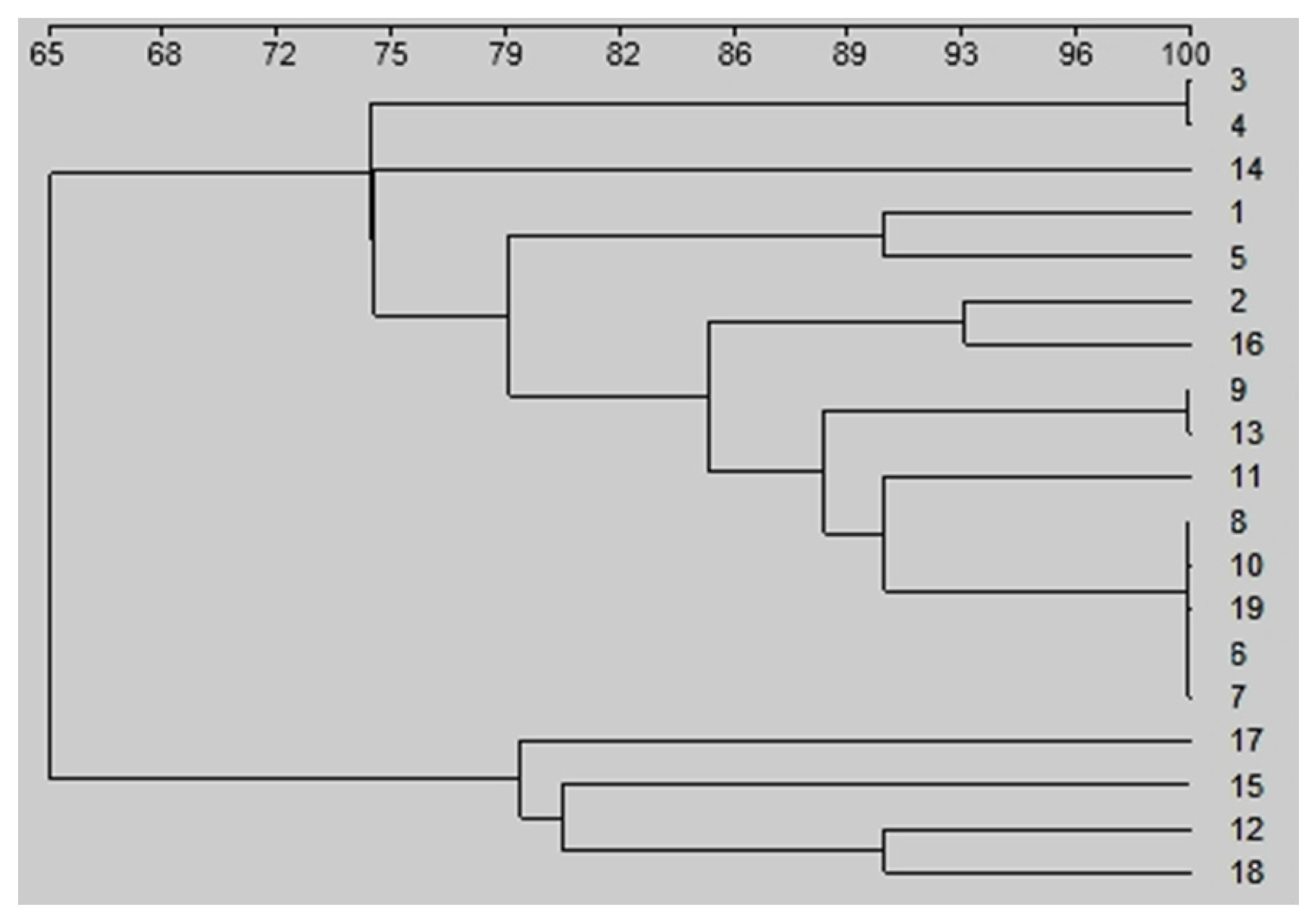
3. Results
4. Discussion
5. Conclusions
Acknowledgements
References
- Debrauwere, H.; Gendrel, C.G.; Lechat, S.; Dutreix, M. Differences and similarities between various tandem repeat sequences: Minisatellites and microsatellites. Biochimie 1997, 79, 577–586. [Google Scholar]
- Chambers, G.K.; MacAvoy, E.S. Microsatellites: Consensus and controversy. Comp. Biochem. Physiol. B 2000, 126, 455–476. [Google Scholar]
- Gur-Arie, R.; Cohen, C.J.; Eitan, Y.; Shelef, L.; Hallerman, E.M.; Kashi, Y. Simple sequence repeats in Escherichia coli: Abundance, distribution, composition, and polymorphism. Genome Res 2000, 10, 62–71. [Google Scholar]
- Tautz, D.; Trick, M.; Dover, G.A. Cryptic simplicity in DNA is a major source of genetic variation. Nature 1986, 322, 652–656. [Google Scholar]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet 2004, 5, 435–445. [Google Scholar]
- Powell, W.; Morgante, M.; Andre, C.; Hanafey, M.; Vogel, J.; Tingey, S.; Rafalski, A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed 1996, 2, 225–238. [Google Scholar]
- Yamamoto, T.; Timura, T.; Sawamura, Y.; Kotobuki, K.; Ban, Y.; Hayashi, T.; Matsuta, N. SSRs isolated from apple can identify polymorphism and genetic diversity in pear. Theor. Appl. Genet 2001, 102, 865–870. [Google Scholar]
- Zane, L.; Bargelloni, L.; Patarnello, T. Strategies for microsatellite isolation: A review. Mol. Ecol 2002, 11, 1–16. [Google Scholar]
- Alexopoulos, C.J.; Mims, C.W.; Blackwell, M. Introductory Mycology, 4th ed; John Wiley & Sons: New York, NY, USA, 1996. [Google Scholar]
- Hawksworth, D.L. The fungal dimension of biodiversity: Magnitude, significance, and conservation. Mycol. Res 1991, 95, 641–655. [Google Scholar]
- Karaoglu, H.; Lee, C.M.Y.; Meyer, W. Survey of simple sequence repeats in completed fungal genomes. Mol. Biol. Evol 2004, 22, 639–649. [Google Scholar]
- Goldstein, D.B.; Pollock, D.D. Launching microsatellites: A review of mutation processes and methods of phylogenetic inference. J. Hered 1997, 88, 335–342. [Google Scholar]
- DeScenzo, R.A.; Harrington, T.C. Use of (CAT) 5 as a DNA fingerprinting probe for fungi. Phytopathology 1994, 84, 534–540. [Google Scholar]
- Barve, M.P.; Haware, M.P.; Sainani, M.N.; Ranjekar, P.K.; Gupta, V.S. Potential of microsatellites to distinguish four races of Fusarium oxysporum f. sp. ciceri prevalent in India. Theor. Appl. Genet 2001, 102, 138–147. [Google Scholar]
- Lasker, J.; Ran, Y. Analysis of Polymorphic Microsatellite Markers for B. A. Typing Penicillium marneffei Isolates. J. Clin. Microbiol. 2004, 42, 1483–1490. [Google Scholar]
- Mwang’Ombe, A.W.; Thiong’O, G.; Olubayo, F.M.; Kiprop, E.K. DNA microsatellite analysis of Kenyan isolates of Rhizoctonis solani from common bean (Phaseolus vulgaris L.). Plant Pathol. J 2007, 6, 66–71. [Google Scholar]
- Alves, A.; Phillips, A.J.L.; Henriques, I.; Correia, A. Rapid differentiation of species of Botryosphaeriaceae by PCR fingerprinting. Res. Microbiol 2007, 158, 112–121. [Google Scholar]
- Wöstemeyer, J.; Kreibich, A. Repetitive DNA elements in fungi (Mycota): Impact on genomic architecture and evolution. Curr. Genet 2002, 41, 189–198. [Google Scholar]
- Geistlinger, J.; Maqbool, S.; Kaiser, W.J.; Kahl, G. Detection of microsatellite fingerprint markers and their Mendelian inheritance in Ascochyta rabei. Mycol. Res 1997, 101, 1113–1121. [Google Scholar]
- Geistlinger, J.; Weising, K.; Kaiser, W.J.; Kahl, G. Allelic variation at a hypervariable compound microsatellite locus in the ascomycete Ascochyta rabiei. Mol. Gen. Genet 1997, 256, 298–305. [Google Scholar]
- Vogelgsang, S.; Enkerli, J.; Jenny, E.; Roffler, S.; Widme, F. 2011 Characterization of Fusarium poae microsatellite markers on strains from Switzerland and other countries. J. Phytopathol 2011, 159, 197–200. [Google Scholar]
- Vogelgsang, S.; Widmer, F.; Jenny, E.; Enkerli, J. Characterisation of novel Fusarium graminearum microsatellite markers in different Fusarium species from various countries. Eur. J. Plant Pathol 2009, 123, 477–482. [Google Scholar]
- Saharan, M.S.; Naef, A. Detection of genetic variation among Indian wheat head scab pathogens (Fusarium spp./isolates) with microsatellite markers. Crop Prot 2008, 27, 1148–1154. [Google Scholar]
- Abd-Elsalam, K.A.; Bahkali, H.A.; Verreet, J.A.; Moslem, M.A. Microsatellite-primed PCR as a tool for discrimination of Fusarium species. J. Food Saf 2011, 31, 35–40. [Google Scholar]
- Balmas, V.; Scherm, B.; di Primo, P.; Rau, D.; Marcello, A.; Migheli, Q. Molecular characterisation of vegetative compatibility groups in Fusarium oxysporum f. sp. radicis-lycopersici and f. sp. lycopersici by random amplification of polymorphic DNA and microsatellite-primed PCR. Eur. J. Plant Pathol. 2005a, 111, 1–8. [Google Scholar]
- Balmas, V.; Scherm, B.; Ghignone, S.; Salem, A.-O.M.; Cacciola, S.O.; Migheli, Q. Characterisation of Phoma tracheiphila by RAPD-PCR, microsatellite-primed PCR and ITS rDNA sequencing and development of specific primers for in planta PCR detection. Eur. J. Plant Pathol. 2005, 111, 235–247. [Google Scholar]
- Lee, S.; Moorman, G.W. Identification and characterization of simple sequence repeat markers for Pythium aphanidermatum, P. cryptoirregulare, and P. irregulare and the potential use in Pythium population genetics. Curr. Genet 2008, 53, 81–93. [Google Scholar]
- Ling, Y.; Zhou, W.; Motohashi, K.; Suga, H.; Fukui, H.; Kageyama, K. Development of microsatellite markers for Pythium helicoides. FEMS Microbiol. Lett 2009, 293, 85–91. [Google Scholar]
- Guillemaut, C.; Edel-Hermann, V.; Camporota, P.; Alabouvette, C.; Richard-Molard, M.; Steinberg, C. Typing of anastomosis groups of Rhizoctonia solani by restriction analysis of ribosomal DNA. Can. J. Microbiol 2003, 49, 556–568. [Google Scholar]
- Abd-Elsalam, K.A.; Guo, J.-R.; Moslem, M.A.; Bahkali, A.H.; Verreet, J.-A. Suitability of intergenic spacer or internal transcribed spacer microsatellite-primed PCR for the identification of phytopathogenic fungi. J. Rapid Autom. Methods Microbiol 2009, 17, 383–397. [Google Scholar]
- Cenis, J.L. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res 1992, 20. [Google Scholar] [CrossRef]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Genetics 2004, 5, 435–445. [Google Scholar]
- Owen, P.G.; Pei, M.; Karp, A.; Royle, D.J.; Edwards, K.J. Isolation and characterization of microsatellite loci in the wheat pathogen Mycosphaerella graminicola. Mol. Ecol 1998, 7, 1611–1612. [Google Scholar]
- Goodwin, S.B.; van der Lee, T.A.J.; Cavaletto, J.R.; Te Lintel Hekkert, B.; Crane, C.F.; Kema, G.H. Identification and genetic mapping of highly polymorphic microsatellite loci from an EST database of the septoria tritici blotch pathogen Mycosphaerella graminicola. Fungal Genet. Biol 2007, 44, 398–414. [Google Scholar]
- Castillo, N.; Cordo, C.; Simón, M. Molecular variability among isolates of Mycosphaerella graminicola, the causal agent of Septoria tritici blotch, in Argentina. Phytoparasitica 2010, 38, 379–389. [Google Scholar]
- Razavi, M.; Hughes, G.R. Microsatellite markers provide evidence for sexual reproduction of Mycosphaerella graminicola in Saskatchewan. Genome 2004, 47, 789–794. [Google Scholar]
- Chen, R.S.; McDonald, B.A. Sexual reproduction plays a major role in the genetic structure of populations of the fungus Mycosphaerella graminicola. Genetics 1996, 142, 1119–1127. [Google Scholar]
- Hoorne, C.; Lamari, J.; Gilbert, J.; Balance, G.M. First report of Mycosphaerella graminicola, the sexual state of Septoria tritici, in Manitoba, Canada. Can. J. Plant Pathol 2002, 24, 445–449. [Google Scholar]
- Singh, R.; Sheoran, S.; Sharma, P.; Chatrath, R. Analysis of simple sequence repeats (SSRs) dynamics in fungus Fusarium graminearum. Bioinformation 2011, 5, 402–404. [Google Scholar]
- Bragança, H.; Rigling, D.; Diogo, E.; Phillips, J.C.A.; Tenreiro, R. Cryphonectria naterciae: A new species in the Cryphonectria–Endothia complex and diagnostic molecular markers based on microsatellite-primed PCR. Fungal Biol 2011, 115, 852–861. [Google Scholar]
| Fungal species | Isolates | Abbrevation | Host | Origin |
|---|---|---|---|---|
| 1. Mycosphaella graminicola | K-Or-1 | Mg | Wheat | Germany |
| 2. | K-Or-30 | Wheat | Kiel, Germany | |
| 3. | K-Or-38 | Wheat | Kiel, Germany | |
| 4. | K-Or-44 | Wheat | Kiel, Germany | |
| 5. | OK-102 | Wheat | Kiel, Germany | |
| 6. | OK-108 | Wheat | Kiel, Germany | |
| 7. | OK-109 | Wheat | Kiel, Germany | |
| 8. | OK-112 | Wheat | Kiel, Germany | |
| 9. | OK-120 | Wheat | Kiel, Germany | |
| 10. | K-Ba-10 | Wheat | Kiel, Germany | |
| 11. | K-Ba-20 | Wheat | Kiel, Germany | |
| 12. | K-Ba-30 | Wheat | Kiel, Germany | |
| 13. | K-Ba-40 | Wheat | Kiel, Germany | |
| 14. | K-Ba-60 | Wheat | Kiel, Germany | |
| 15. | G-Or-1 | Wheat | Kiel, Germany | |
| 16. | G-Or-6 | Wheat | Göttingen, Germany | |
| 17. | G-Or-8 | Wheat | Göttingen, Germany | |
| 18. | G-Or-88 | Wheat | Göttingen, Germany | |
| 19. | G-Or-102 | Wheat | Göttingen;Germany | |
| Fusarium oxysporum f. sp. vasinfectum | Fov | Cotton | Egypt | |
| F. solani | Fs | Cotton | Egypt | |
| F. germanium | Isolate 37 | Fg | Wheat | Germany |
| F. culmorum | Fcu | Wheat | Germany | |
| F. poae | Fp | Wheat | Germany | |
| Macrophomina phaseolina | Mp | Cotton | Egypt | |
| Trichoderma harizinum | Th | Cotton | Egypt | |
| Septoria tritici | St | Wheat | Germany | |
| Stagonospora nodorum | GB1 | Sn | Wheat | Germany |
| Pyrenophora teres | Pte | Barley | Germany | |
| Pyrenophora tritici-repentis. | Greifenhagen I | Ptr | Wheat | Germany |
| Pseudocercosporella herpotrichoides | Ph | Wheat | Germany | |
| Penicillium sp. | Pen | Wheat | Germany | |
| Alternaria sp. | Alt | Wheat | Germany | |
| Cercospora beticola | Cb | Sugerbeet | Germany | |
| Chaetomium sp. | Ch | Cotton | Egypt |
| Nucleotides repeats | Primers | Motif | Length | GC Content (%) | MW | Optimum AT (°C) | Range of fragment size (bp) |
|---|---|---|---|---|---|---|---|
| Di | (CT)7G | CT | 15 | 53.3 | 4420.9 | 40 | 400–2000 |
| (CA)7T | CA | 15 | 46.7 | 4458.9 | 52 | 300–1700 | |
| (AG)7C | AG | 15 | 53.3 | 4724.1 | 48 | 200–2500 | |
| Tri | (CTG)5 | CTG | 15 | 66.7 | 4550.9 | 52 | 450–2000 |
| (AGG)5 | AGG | 15 | 66.7 | 4796.1 | 52 | 350–2000 | |
| (TCC)5 | TCC | 15 | 66.7 | 4350.8 | 50 | 400–2000 | |
| (ACG)5 | ACG | 15 | 66.7 | 4596.0 | 50 | 400–2000 | |
| (TGG)5 | TGG | 15 | 66.7 | 4751.1 | 50 | 350–1500 | |
| (GTA)5 | GTA | 15 | 33.3 | 4671.1 | 40 | 600–2000 | |
| (ATG)5 | ATG | 15 | 33.3 | 4671.1 | 50 | 200–3000 | |
| (TAC)5 | TAC | 15 | 33.3 | 4470.9 | 40 | 700–1500 | |
| (TGC)5 | TGC | 15 | 66.7 | 4550.9 | 40 | 1000–1800 | |
| (GCT)5 | GCT | 15 | 66.7 | 4550.9 | 52 | 600–2000 | |
| (TGT)5 | TGT | 15 | 33.3 | 4626.0 | 40 | 400–1800 | |
| Tetra | (TGAC)4 | TGAC | 16 | 50.0 | 4881.2 | 55 | 400–2500 |
| (GTCA)4 | GTCA | 16 | 50.0 | 4881.2 | 50 | 400–1800 | |
| (TAGG)4 | TAGG | 16 | 50.0 | 5041.3 | 50 | 200–2000 | |
| (TTTC)4 | TTTC | 16 | 25.0 | 4745.1 | 40 | 300–2000 | |
| (TACC)4 | TACC | 16 | 50.0 | 4721.1 | 45 | 550–2000 | |
| (GGTT)4 | GGTT | 16 | 50.0 | 5005.3 | 40 | 550–2500 | |
| MP Primers | Number of alleles amplified from different fungal species | Total No. of Band | Mean alleles amplified per species | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fov | Fs | Fg | Fcu | Fp | Mp | Th | St | Sn | Pte | Ptri | Ph | Pen | Alt | Cb | Ch | |||
| (CT)7G | 2 | 4 | 4 | 3 | 5 | 0 | 1 | 3 | 5 | 3 | 3 | 2 | 4 | 2 | 0 | 3 | 43 | 2.7 |
| (CA)7T | 4 | 3 | 7 | 4 | 23 | 5 | 3 | 3 | 4 | 3 | 2 | 3 | 4 | 4 | 5 | 4 | 61 | 3.8 |
| (AG)7C | 1 | 2 | 2 | 2 | 3 | 4 | 3 | 6 | 0 | 4 | 6 | 2 | 4 | 8 | 3 | 5 | 52 | 3.3 |
| (CTG)5 | 4 | 4 | 4 | 4 | 5 | 4 | 4 | 5 | 4 | 6 | 4 | 4 | 5 | 4 | 7 | 3 | 71 | 4.4 |
| (AGG)5 | 10 | 8 | 8 | 7 | 8 | 5 | 6 | 4 | 8 | 6 | 4 | 6 | 5 | 5 | 6 | 7 | 103 | 6.4 |
| (TCC)5 | 8 | 8 | 9 | 9 | 4 | 5 | 4 | 6 | 5 | 4 | 6 | 5 | 6 | 5 | 3 | 3 | 90 | 5.6 |
| (ACG)5 | 6 | 6 | 4 | 6 | 7 | 6 | 5 | 7 | 10 | 7 | 8 | 7 | 7 | 7 | 7 | 7 | 107 | 6.7 |
| (TGG)5 | 3 | 5 | 5 | 5 | 7 | 6 | 4 | 6 | 7 | 5 | 7 | 5 | 6 | 6 | 6 | 8 | 91 | 5.7 |
| (GTA)5 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 3 | 0 | 3 | 3 | 0 | 3 | 0 | 3 | 0 | 17 | 1.1 |
| (ATG)5 | 4 | 5 | 6 | 2 | 7 | 3 | 4 | 5 | 1 | 3 | 4 | 4 | 5 | 2 | 4 | 4 | 63 | 3.9 |
| (TAC)5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0.2 |
| (TGC)5 | 3 | 3 | 1 | 2 | 3 | 0 | 0 | 3 | 2 | 5 | 1 | 3 | 3 | 0 | 3 | 0 | 32 | 2.0 |
| (GCT)5 | 3 | 2 | 4 | 3 | 3 | 0 | 1 | 2 | 1 | 2 | 3 | 3 | 2 | 2 | 4 | 3 | 38 | 2.4 |
| (TGT)5 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 1 | 0 | 9 | 0.6 |
| (TGAC)4 | 6 | 3 | 6 | 4 | 6 | 4 | 1 | 1 | 6 | 5 | 4 | 4 | 5 | 8 | 2 | 2 | 67 | 4.2 |
| (GTCA)4 | 1 | 2 | 4 | 5 | 2 | 4 | 2 | 4 | 3 | 2 | 2 | 5 | 3 | 2 | 2 | 1 | 44 | 2.8 |
| (TAGG)4 | 4 | 4 | 8 | 3 | 4 | 3 | 2 | 3 | 4 | 3 | 5 | 3 | 5 | 3 | 5 | 3 | 62 | 3.8 |
| (TTTC)4 | 3 | 1 | 3 | 1 | 3 | 2 | 1 | 2 | 4 | 3 | 2 | 2 | 4 | 2 | 2 | 3 | 38 | 2.4 |
| (TACC)4 | 5 | 4 | 3 | 2 | 4 | 0 | 0 | 4 | 4 | 1 | 2 | 3 | 4 | 7 | 2 | 3 | 48 | 3.0 |
| (GGTT)4 | 1 | 3 | 1 | 1 | 4 | 0 | 0 | 4 | 5 | 2 | 2 | 2 | 2 | 5 | 1 | 3 | 36 | 2.3 |
| T3B | 5 | 5 | 7 | 7 | 10 | 6 | 7 | 8 | 9 | 7 | 6 | 5 | 4 | 5 | 4 | 6 | 105 | 6.6 |
| M13 | 4 | 4 | 6 | 5 | 4 | 4 | 2 | 2 | 6 | 4 | 4 | 3 | 5 | 7 | 5 | 4 | 69 | 4.3 |
| (GTG)5 | 5 | 6 | 6 | 5 | 8 | 4 | 6 | 4 | 4 | 4 | 4 | 6 | 8 | 8 | 8 | 4 | 90 | 5.7 |
| (GTGC)4 | 6 | 5 | 3 | 6 | 2 | 4 | 4 | 2 | 1 | 3 | 4 | 5 | 6 | 4 | 4 | 3 | 62 | 3.9 |
| (CAG)3 | 4 | 5 | 5 | 4 | 5 | 5 | 5 | 6 | 4 | 4 | 5 | 5 | 4 | 5 | 4 | 5 | 75 | 4.7 |
| Primers | Total bands | Polymorphic bands | PB% | ||||
|---|---|---|---|---|---|---|---|
| Fg | Fcu | Total | Fg | Fcu | Total | ||
| (CT)7G | 4 | 3 | 7 | 2 | 1 | 3 | 42.9 |
| (CA)7 T | 7 | 4 | 11 | 4 | 1 | 5 | 45.5 |
| (AG)7C | 2 | 2 | 4 | 2 | 2 | 4 | 100.0 |
| (CTG)5 | 4 | 4 | 8 | 2 | 2 | 4 | 50.0 |
| (AGG)5 | 8 | 7 | 15 | 2 | 2 | 4 | 26.7 |
| (TCC)5 | 9 | 9 | 18 | 4 | 3 | 7 | 38.9 |
| (ACG)5 | 4 | 6 | 10 | 0 | 2 | 2 | 33.3 |
| (TGG)5 | 5 | 5 | 10 | 0 | 0 | 0 | 0 |
| (GTA)5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| (ATG)5 | 6 | 2 | 8 | 3 | 0 | 3 | 37.5 |
| (TAC)5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| (TGC)5 | 1 | 2 | 3 | 1 | 1 | 2 | 67.7 |
| (GCT)5 | 4 | 3 | 7 | 2 | 0 | 2 | 28.6 |
| (TGT)5 | 1 | 0 | 1 | 1 | 0 | 1 | 100.0 |
| (TGAC)4 | 6 | 4 | 10 | 3 | 1 | 4 | 40.0 |
| (GTCA)4 | 4 | 5 | 9 | 2 | 2 | 4 | 44.4 |
| (TAGG)4 | 8 | 3 | 11 | 6 | 2 | 8 | 72.7 |
| (TTTC)4 | 3 | 1 | 4 | 3 | 1 | 4 | 100 |
| (TACC)4 | 3 | 2 | 5 | 2 | 1 | 1 | 40.0 |
| (GGTT)4 | 1 | 1 | 2 | 0 | 0 | 0 | 0 |
| *T3B | 7 | 7 | 14 | 2 | 2 | 4 | 28.6 |
| *M13 | 6 | 5 | 11 | 1 | 1 | 2 | 18.2 |
| (GTG)5 | 6 | 5 | 11 | 1 | 0 | 1 | 9.09 |
| (GTGC)4 | 3 | 6 | 9 | 1 | 4 | 5 | 55.6 |
| (CAG)3 | 5 | 4 | 9 | 2 | 0 | 2 | 22.2 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Bahkali, A.H.; Abd-Elsalam, K.A.; Guo, J.-R.; Khiyami, M.A.; Verreet, J.-A. Characterization of Novel Di-, Tri-, and Tetranucleotide Microsatellite Primers Suitable for Genotyping Various Plant Pathogenic Fungi with Special Emphasis on Fusaria and Mycospherella graminicola. Int. J. Mol. Sci. 2012, 13, 2951-2964. https://doi.org/10.3390/ijms13032951
Bahkali AH, Abd-Elsalam KA, Guo J-R, Khiyami MA, Verreet J-A. Characterization of Novel Di-, Tri-, and Tetranucleotide Microsatellite Primers Suitable for Genotyping Various Plant Pathogenic Fungi with Special Emphasis on Fusaria and Mycospherella graminicola. International Journal of Molecular Sciences. 2012; 13(3):2951-2964. https://doi.org/10.3390/ijms13032951
Chicago/Turabian StyleBahkali, Ali H., Kamel A. Abd-Elsalam, Jian-Rong Guo, Mohamed A. Khiyami, and Joseph-Alexander Verreet. 2012. "Characterization of Novel Di-, Tri-, and Tetranucleotide Microsatellite Primers Suitable for Genotyping Various Plant Pathogenic Fungi with Special Emphasis on Fusaria and Mycospherella graminicola" International Journal of Molecular Sciences 13, no. 3: 2951-2964. https://doi.org/10.3390/ijms13032951





